READY TO STEP INTO THE NEXT LEVEL OF HLA SEQUENCING?
Contact us here and we will provide you all the necessary information to start NanoTYPing with Omixon.
ABOUT NANOTYPE
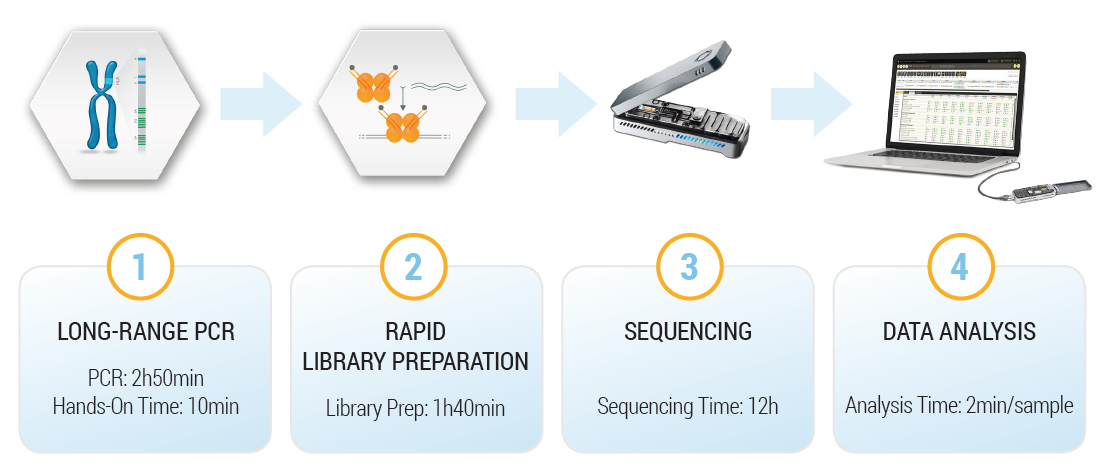
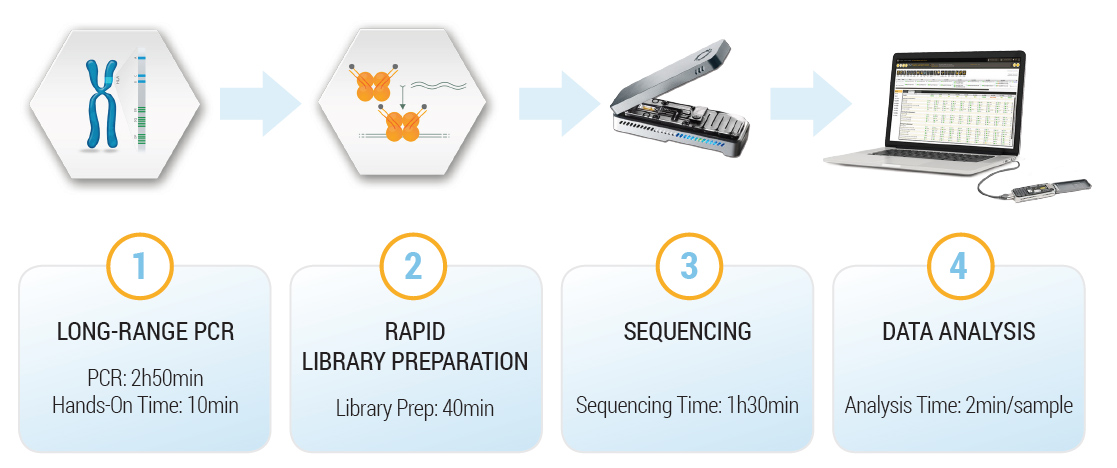
NanoTYPE™ is a HLA typing assay using third generation sequencing technology from Oxford Nanopore Technologies (ONT). The kit contains all necessary reagents for the amplification of your sample and comes with our tailored genotyping software, NanoTYPER™. The kit has been designed to be combined with a rapid barcoding library preparation from ONT.
All 11 loci (HLA-A, B, C, DQA1, DQB1, DPA1, DPB1, DRB1 and DRB345) are amplified in a single long-range multiplex PCR. The user can choose between 2 different protocols, either for a single sample urgent typing or multiple samples.
The easy workflow, the rapid turnaround time, the long reads and the absence of capital investment for the instrument makes it an ideal primary or a complementary technique in your lab.
About Nanopore-Based Typing
Oxford Nanopore has developed a new generation of sensing technology that uses nanopores embedded in high tech electronics to perform precise molecular analysis. When a DNA fragment is entering a nanopore, each DNA base is disrupting the electrical field with a specific signature and can be used as a single molecule detector. The deconvolution of the electrical signal is done using a basecaller converting the electric signal into a DNA sequence with a FASTQ output format This FASTQ file is then imported into NanoTYPER software for genotyping. One major advantage of nanopore sequencing is the long reads facilitating the phasing of even long-distant SNPs resulting in lower number of ambiguities.
“This technology may represent a paradigm shift similar to Luminex Technology for Antibody assessment in the early / mid 2000’s”
*average accuracy based on 228 samples with 2 field references for all loci
EXPERTS’ FEEDBACK

“The technology is cost effective, scalable for the parallel typing of one-to-many samples, in a short period of time, providing excellent opportunities for the typing of deceased donors, elevating our matching capabilities to allow for epitope matching of these donors.”
PROF. DIMITRI S. MONOS, PhD
Pathology and Laboratory Medicine
Perelman School of Medicine
University of Pennsylvania
The Children’s Hospital of Philadelphia, USA
“We were astonished by the potential impact of this solution to enable sensitized patients to benefit from reliable epitope-based matching of deceased donors for the first time.”
PROF. JEAN-LUC TAUPIN, PharmD, PhD
President of the Francophone Society of Histocompatibility and Immunogenetics
Immunology and Histocompatibility Laboratory, Saint-Louis Hospital, AP-HP, Paris, France
“Overall, Nanopore sequencing is an excellent option for the future of HLA typing methodology.“
DR. ANA M LAZARO-SHIBEN, PhD, CHS
Immunogenetics Laboratory
Johns Hopkins University School of
Medicine, Baltimore, USA
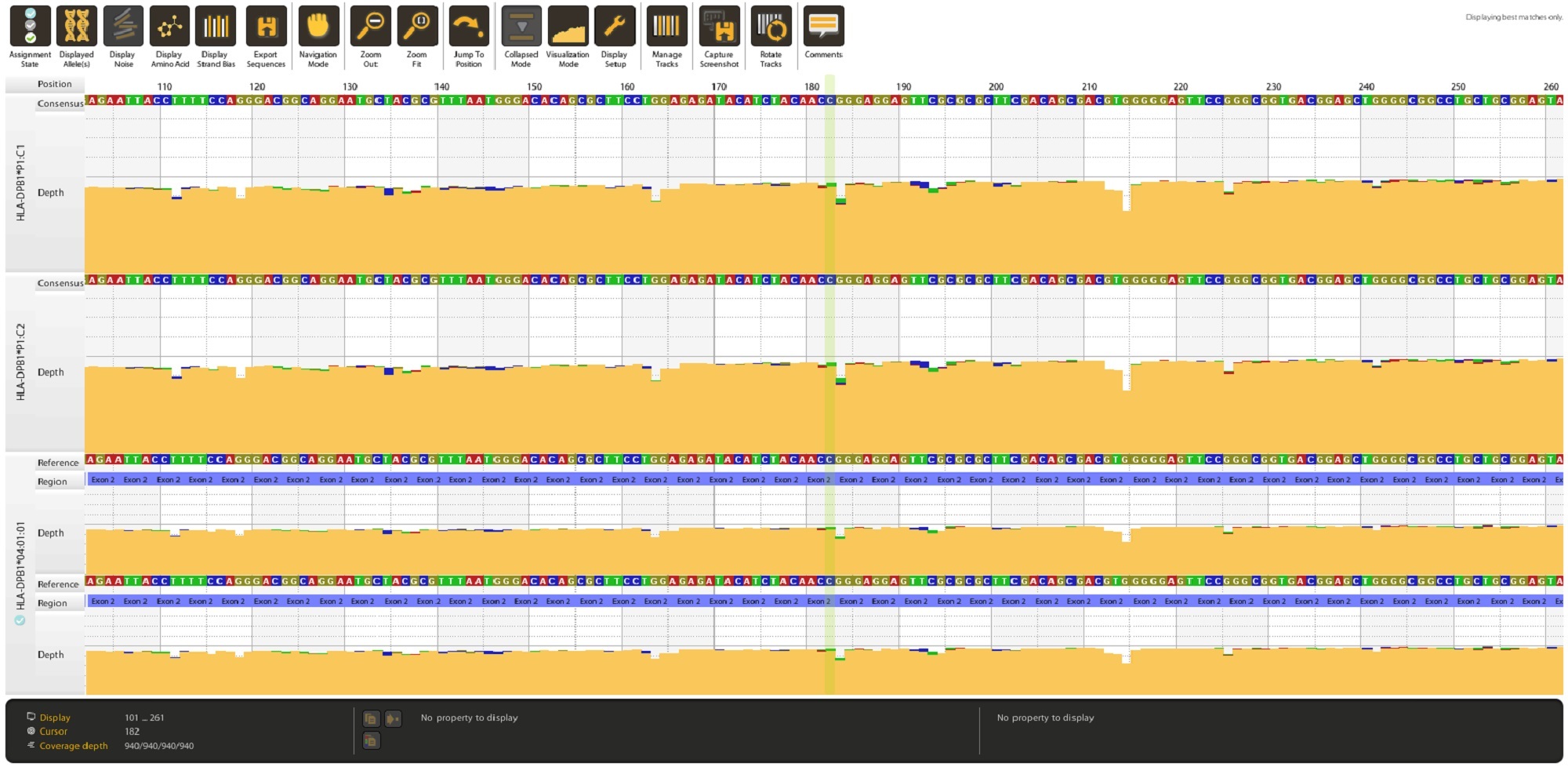
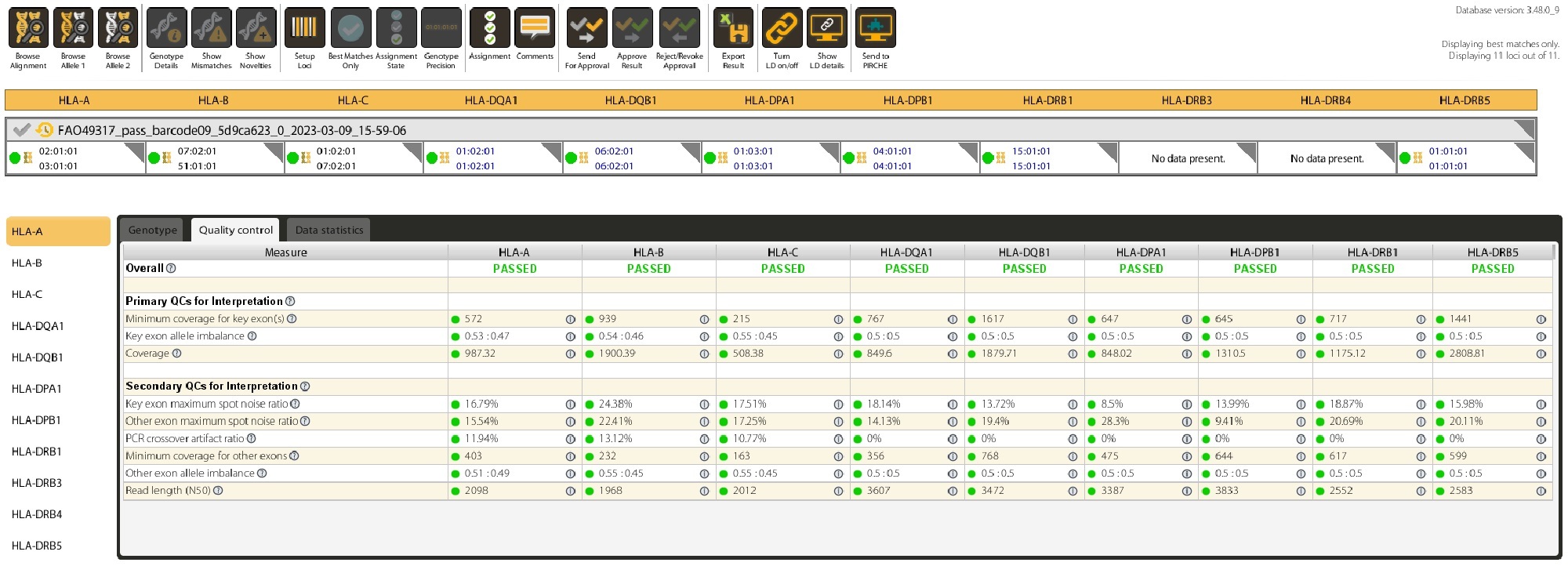
NANOTYPER SOFTWARE ANALYSIS
Long-read sequences are analyzed using our intuitive and proprietary NanoTYPER software.
The quality of the results is assessed using various metrics to aid in accepting or rejecting your results.
Data are exported from NanoTYPER software as JSON, PDF, HML, CSV, XML, TXT format and can be integrated into your LIM system.
READY TO STEP INTO THE NEXT LEVEL OF HLA SEQUENCING?
Contact us here and we will provide you all the necessary information to start NanoTYPing with Omixon.
MinION® is a registered trademark of Oxford Nanopore Technologies. NanoTYPE and NanoTYPER are trademarks of Omixon Biocomputing Ltd.