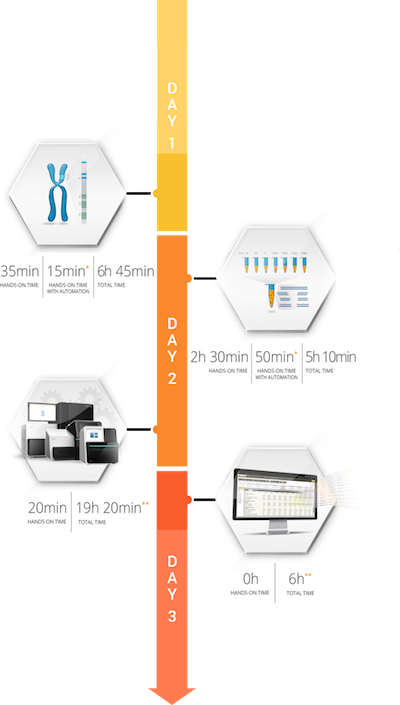
Sample Preparation
- Less than 0.8µg of DNA required for 11 locus amplification
- Compatible with multiple DNA sources: whole blood, cord blood, buccal swabs & saliva
- 5, 7 or 11 locus amplifications
- Scalable up to 96 samples / 11 loci, up to 192 samples / 7 loci and 288 / 5 loci***
Library Preparation
- 4 hours hands-on time for 24 samples
- Less than 48 hours turnaround time from sample to result
- Library Size Selection: Pippin Prep or Magnetic Beads
- Holotype libraries can be combined with other libraries for optimized sequencing cost
Data Analysis with HLA Twin
- Fully automated analysis after Illumina sequencing run
- Less than 5 minutes analysis time per 11 loci sample with recommended specifications
- Two independent genotyping algorithms
- 24 Quality Control Metrics for confident assignment
- Balanced coverage between alleles at a locus, between loci in a sample and among samples
- Deep and even coverage across the whole length of the targeted region
All timings are for 24 sample, Holotype HLA v3.0
* Timings are based on the liquid handler used
** On a MiSeq 300-cycle Micro flow cell
*** On a 300-cycle Standard flow cell with cluster density ~1,000K/mm²