Confidence
- Two independent algorithms - Consensus Genotyping and Statistical Genotyping
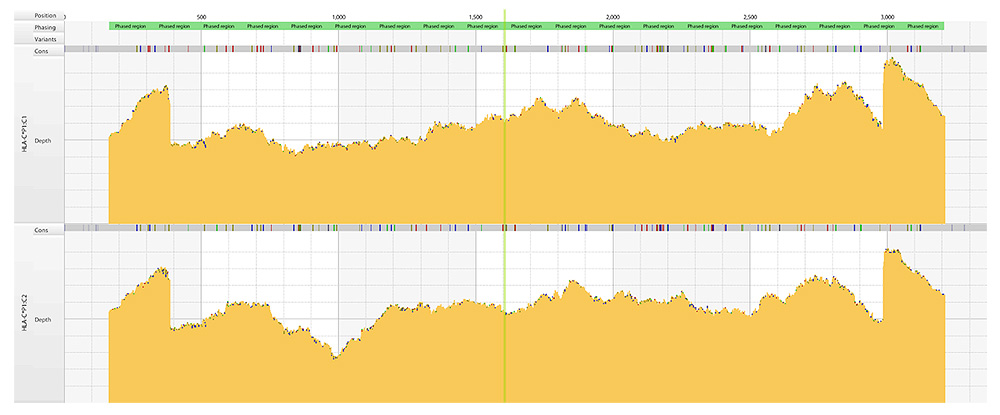
- Consensus Genotyping:
- De novo assembly: optimal for fully phased consensus sequences
- Reference alignment: added analytical confidence, novel allele detection and null allele resolution
- Analyze data against any version of the IMGT/HLA Database
- DRB, DQ Linkage Disequilibrium checks
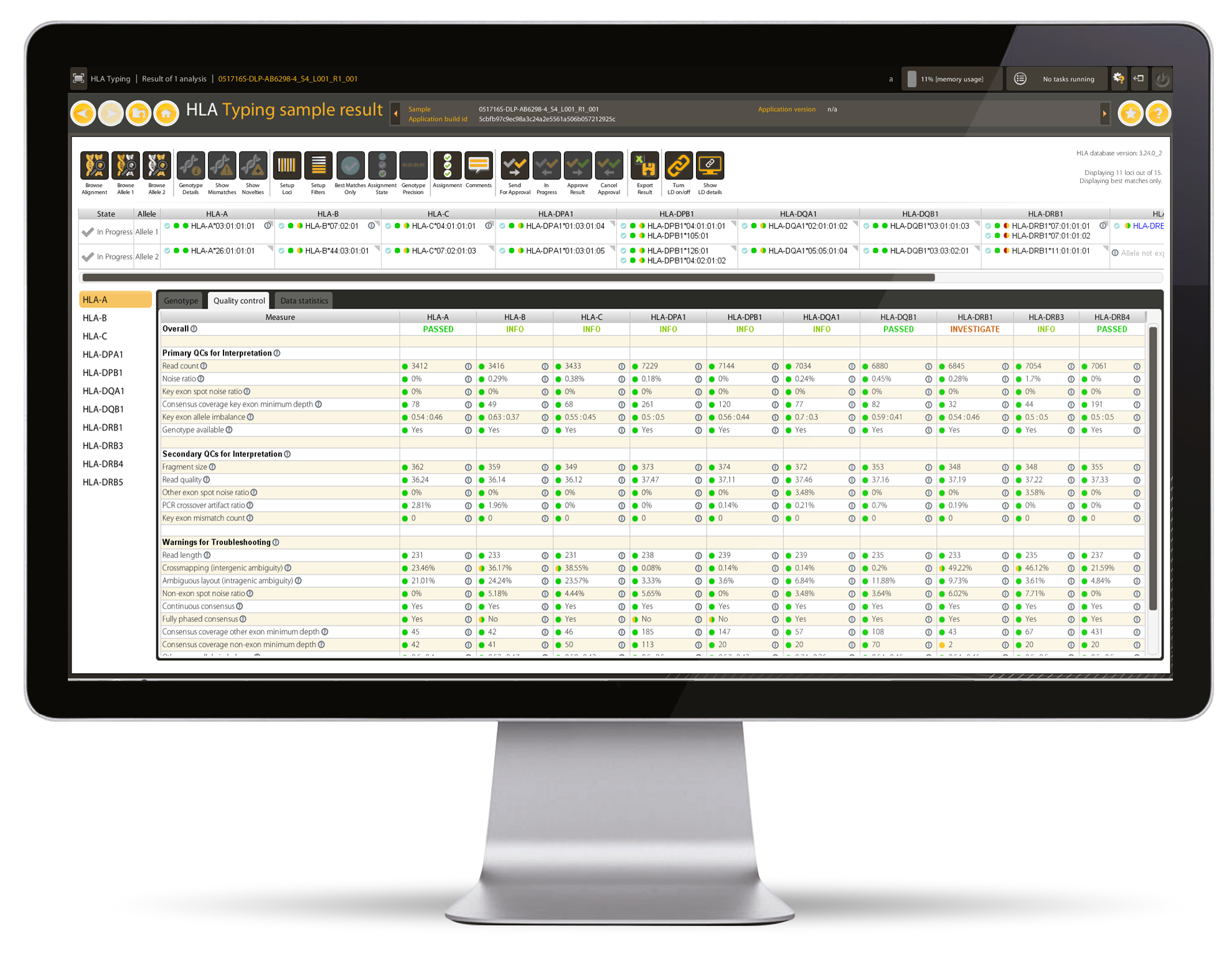
Quality
- 24 Quality Control metrics for confident assignment and detailed troubleshooting
- Traffic Light System for easy interpretation of genotyping results
- Whole Gene Consensus sequences for unambiguous allele assignment
- Complete Novel and Null allele detection
Automation and Integration
- Fully automated analysis after Illumina sequencing run
- Export genotyping results - HML, PDF, JSON, XLSX, CSV, TXT and HPRIM
- Integration with LIMS, such as Histotrac and mTilda
- Simple assignment and multi-layer approval workflows
- Simple and customizable allele assignment for ease of use
Flexibility & Scalability
- Platform independent (available for Windows, Linux and macOS)
- Desktop and Client-Server versions
- Customizable analysis and visualization