Authors: Yuxin Yin, James H. Lan, David Nguyen, Nicole Valenzuela, Ping Takemura, Yung-Tsi Bolon, Brianna Springer, Katsuyuki Saito, Ying Zheng, Tim Hague, Agnes Pasztor, Gyorgy Horvath, Krisztina Rigo, Elaine F. Reed, Qiuheng Zhang
Abstract:
Background
Unambiguous HLA typing is important in hematopoietic stem cell transplantation (HSCT), HLA disease association studies, and solid organ transplantation. However, current molecular typing methods only interrogate the antigen recognition site (ARS) of HLA genes, resulting in many cis-trans ambiguities that require additional typing methods to resolve. Here we report high-resolution HLA typing of 10,063 National Marrow Donor Program (NMDP) registry donors using long-range PCR by next generation sequencing (NGS) approach on buccal swab DNA.
Methods
Multiplex long-range PCR primers amplified the full-length of HLA class I genes (A, B, C) from promotor to 3’ UTR. Class II genes (DRB1, DQB1) were amplified from exon 2 through part of exon 4. PCR amplicons were pooled and sheared using Covaris fragmentation. Library preparation was performed using the Illumina TruSeq Nano kit on the Beckman FX automated platform. Each sample was tagged with a unique barcode, followed by 2×250 bp paired-end sequencing on the Illumina MiSeq. HLA typing was assigned using Omixon Twin software that combines two independent computational algorithms to ensure high confidence in allele calling. Consensus sequence and typing results were reported in Histoimmunogenetics Markup Language (HML) format. All homozygous alleles were confirmed by Luminex SSO typing and exon novelties were confirmed by Sanger sequencing.
Results
Using this automated workflow, over 10,063 NMDP registry donors were successfully typed under high-resolution by NGS. Despite known challenges of nucleic acid degradation and low DNA concentration commonly associated with buccal-based specimens, 97.8% of samples were successfully amplified using long-range PCR. Among these, 98.2% were successfully reported by NGS, with an accuracy rate of 99.84% in an independent blind Quality Control audit performed by the NDMP. In this study, NGS-HLA typing identified 23 null alleles (0.023%), 92 rare alleles (0.091%) and 42 exon novelties (0.042%).
Conclusion
Long-range, unambiguous HLA genotyping is achievable on clinical buccal swab-extracted DNA. Importantly, full-length gene sequencing and the ability to curate full sequence data will permit future interrogation of the impact of introns, expanded exons, and other gene regulatory sequences on clinical outcomes in transplantation.
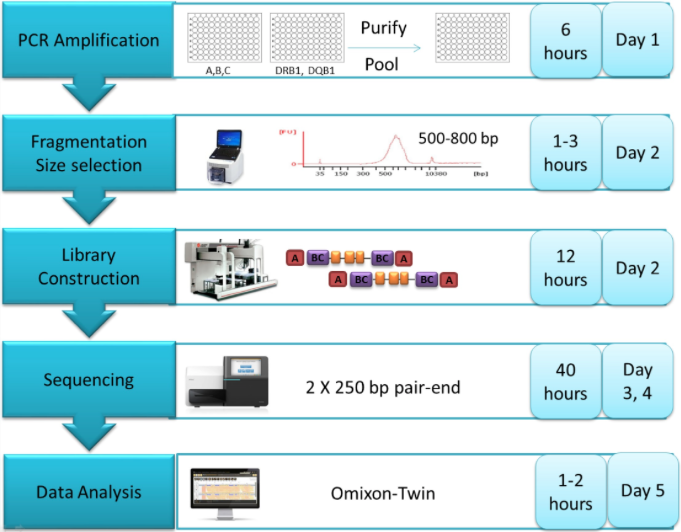
High-throughput NGS workflow begins with multiplex long range PCR of A, B, C and DRB1, DQB1. After PCR, amplicons are purified and pooled in equimolar concentrations. Sheared amplicons then undergo library preparation by using the Illumina TruSeq Nano Kit. To maximize throughput, each clinical sample is labeled with unique dual indices. 2×250 bp paired-end sequence data from the Illumina MiSeq are exported and analyzed using Omixon Twin1.0.7, with 3.19.0 IMGT/HLA database serving as the reference.
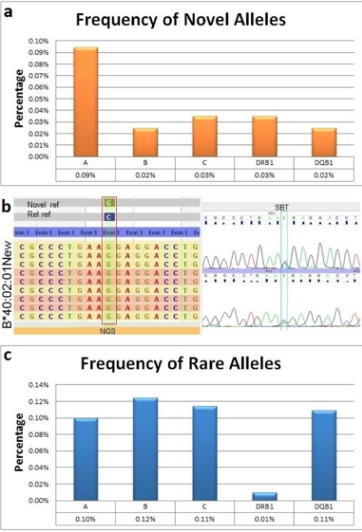
Sequence data was analyzed using Omixon HLA Twin V1.0.7 (3.19.0 IMGT/HLA database). a) Percentage of SBT-confirmed exon novelties shown by HLA locus. b) Example of a novel allele detected by NGS and confirmed using SBT. SBT was unable to determine the cis-trans phase of the exon novelty; in contrast, parallel sequencing by NGS clearly established the phase and location of the novel variant in the B*40:02:01 new allele. c) Percentage of rare alleles detected shown by HLA locus.
For all figures and tables, read the original paper on PLOS ONE…






