Written by Vera Siffnerová (FAS, Omixon)
A choice of the most suitable donor for each transplant patient has always been the main focus and challenge for the whole team that is involved in the selection process. There is a high number of criteria that are evaluated, different approaches and the needs to be considered throughout this process are usually highly standardized across different transplant centres. However, some of them can vary based on the patient’s diagnosis, unique experiences of each transplant centre and can depend on having access to improved methods as well as the attitude to experiments with novel approaches for cure.
Continuous research and development of immunology together with medical treatment evolution has a major positive impact on general transplant outcome. Therefore, the decision of selecting the best donor was not getting easier with many different approaches evolving when different strategies can be applied. As an example, we can review one of the criteria, which has been in focus nowadays (besides the traditional HLA-based matching) and that is HLA mismatching based on the growing knowledge on Natural Killer (NK) cells, which are strongly involved in the transplantation success.
NK cells are a type of cytotoxic lymphocytes that have an important role in innate immunity. They act as the first line of defense against viral infections, early cellular transformation and tumor growth. The “missing-self” concept, put forward in the 1980s, formed the basis for understanding NK-derived allorecognition by describing a mechanism by which NK cells target tumor cells deficient in Major Histocompatibility Complex (MHC) Class I expression. These cells are equipped with a number of surface receptors, inhibitory, activating, or both, that control their activity. A major group of these regulatory receptors is called the Killer-cell Immunoglobulin-like Receptors (KIRs) and have been studied in numerous diseases. They are genetically the most polymorphic of these receptors, interacting with the equally polymorphic HLA class I system (1-3).
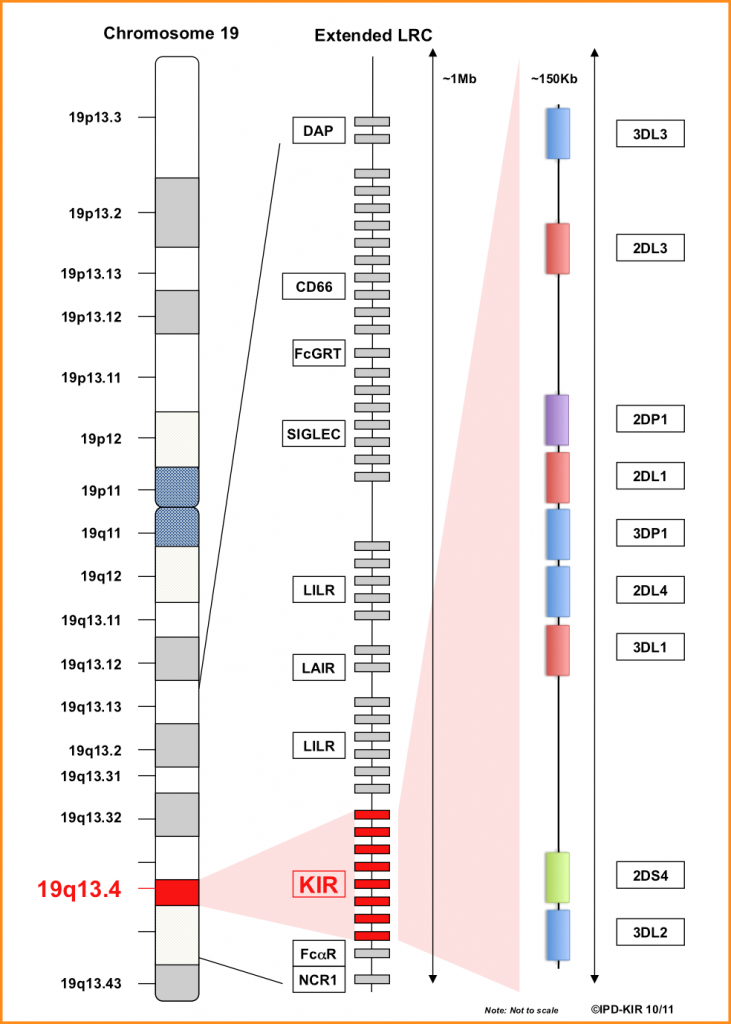
KIR molecules are encoded within a 100-200 Kb region of the Leukocyte Receptor Complex (LRC) present on chromosome 19 (19q13.4) which is segregated independently of the HLA genes. LRC is polygenic, individual genes exhibit polymorphism and between them 15 KIR genes (KIR2DL1, KIR2DL2/L3, KIR2DL4, KIR2DL5A, KIR2DL5B, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, KIR3DL1/S1, KIR3DL2, KIR3DL3) and two KIR pseudogenes (KIR2DP1 and KIR3DP1) are officially recognized (Fig 1) (4,5).
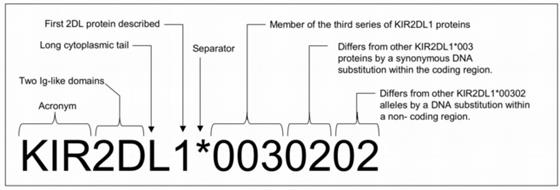
Today the human KIR family is represented by 1110 alleles (listed in the IPD-KIR Database v2.9.0, December 2019). Three criteria – number of extracellular Ig-like domains (2D/3D), cytoplasmic tail length (L/S), and sequence similarity (number code) – have been used to classify the encoded KIR proteins into different groups and used for the KIR nomenclature (Fig 2, Fig 4) (6,7).
Figure 2: an example of KIR allele nomenclature
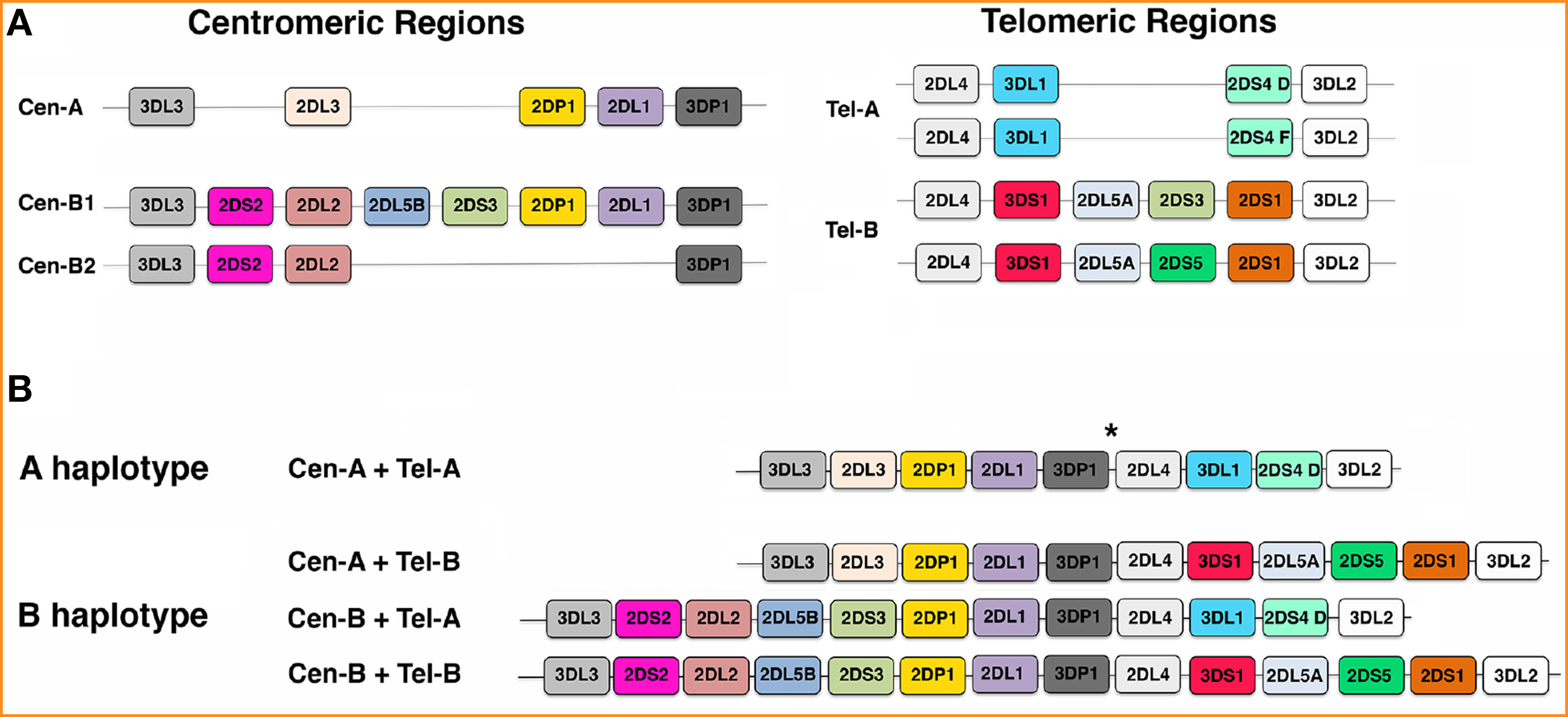
KIR genes are organised into two basic groups based on the gene content – haplotype A and haplotype B. They show an extensive variation in the number and type of KIR genes present. Haplotype A, which is the most common one, is usually inhibitory as it contains 6 inhibitory genes: KIR2DL1, KIR2DL3 (most characteristic), KIR3DL1, KIR3DL2, KIR3DL3 and KIR2DL4 and one activating gene: KIR2DS4. These genes are more frequently found in haplotype A than B but are not exclusive to it and also have a substantial allelic diversity. Haplotype B, on the other hand, has a very diverse gene content in addition to minor allelic diversity. It is mostly activating and has a variable number of activating KIRs (ranging from 2 to 7), and a similar number of inhibitory KIRs as haplotype A. Certain genes, however, occur exclusively on haplotype B such as: KIR2DS1, KIR2DS2-2DL2 (most characteristic), KIR2DS3, KIR2DS5, KIR2DL2, KIR2DL5 and KIR3DS1. So far, 27 different KIR haplotypes have been described (http://www.imgt.org/) (8).
With few exceptions, KIR haplotypes can be divided in two regions, centromeric (Cen) and telomeric (Tel), by the presence of a recombination hotspot. Four genes, named framework genes (KIR3DL3, KIR3DP1, KIR2DL4, and KIR3DL2), mark the beginning and the end of these two regions, and which, with very few exceptions, are present in every individual (Fig 1). Based on the different number and kind of KIR genes present, three different centromeric regions (referred to as Cen-A, Cen-B1, and Cen-B2) and two different telomeric regions (namely Tel-A and Tel-B) have been identified (Fig 3) (9).
Figure 3: Gene organization of KIR locus. (A) A representation of the most frequent centromeric (Cen) and telomeric (Tel) regions detected in Caucasians. (B) Schematic pictures of KIR gene order in the A haplotypes and in 3 representative B haplotypes. * indicates the hot spot of recombination.
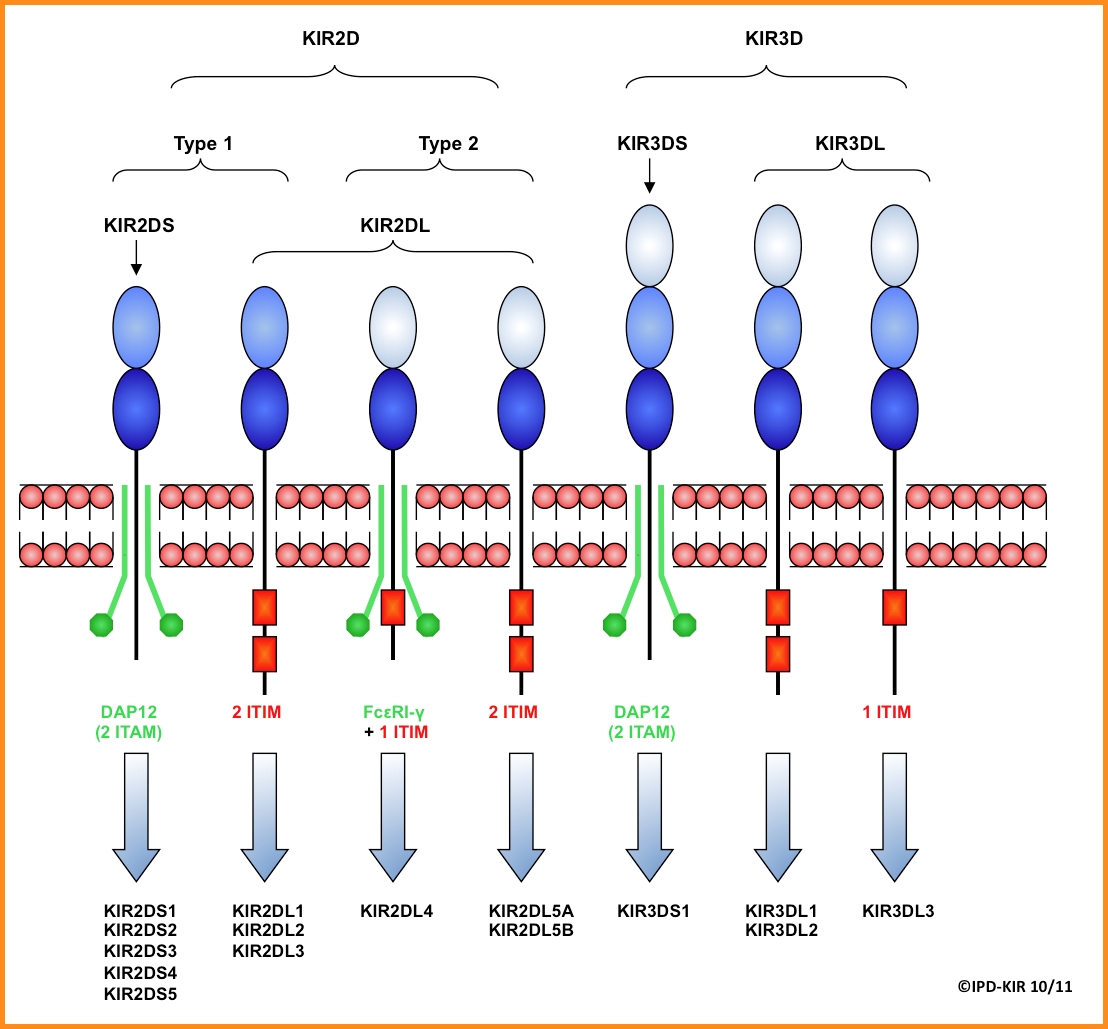
KIR proteins consist of characteristic Ig-like domains on their extracellular regions, which some of them are involved in HLA class I ligand binding. They also possess transmembrane and cytoplasmic regions which are functionally relevant as they define the type of signal which is transduced to the NK cell. KIR proteins can have two or three Ig-like domains (hence KIR2D or KIR3D) as well as short or long cytoplasmic tails (represented as KIR2DS or KIR2DL). Two domain KIR proteins are subdivided into two groups depending on the origin of the membrane distal Ig-like domains present. Long cytoplasmic tails usually contain two Immune Tyrosine-based Inhibitory Motifs (ITIM) which transduce inhibitory signals to the NK cell (inhibitory KIRs, iKIRs). Short cytoplasmic tails possess a positively charged amino acid residue in their transmembrane region which allows them to associate with a DAP12 signalling molecule capable of generating an activation signal (activating KIRs, aKIRs) (6,9).
Figure 4: KIR protein structures. The structural characteristics of two and three Ig-like domain KIR proteins are shown. The association of activating KIR to adaptor molecules is shown in green, whereas the ITIM of inhibitory KIR are shown as red boxes.
Further variation in KIR repertoires between individuals results from the relatively polymorphic nature of the KIR genes and expression differences can occur due to null/low/high expression allele variants and copy number variation. Moreover, a salient feature of KIRs is their stochastic distribution of cell-surface expression across the NK cell compartment, resulting in highly diverse NK cell KIR repertoires unique for each individual and population.
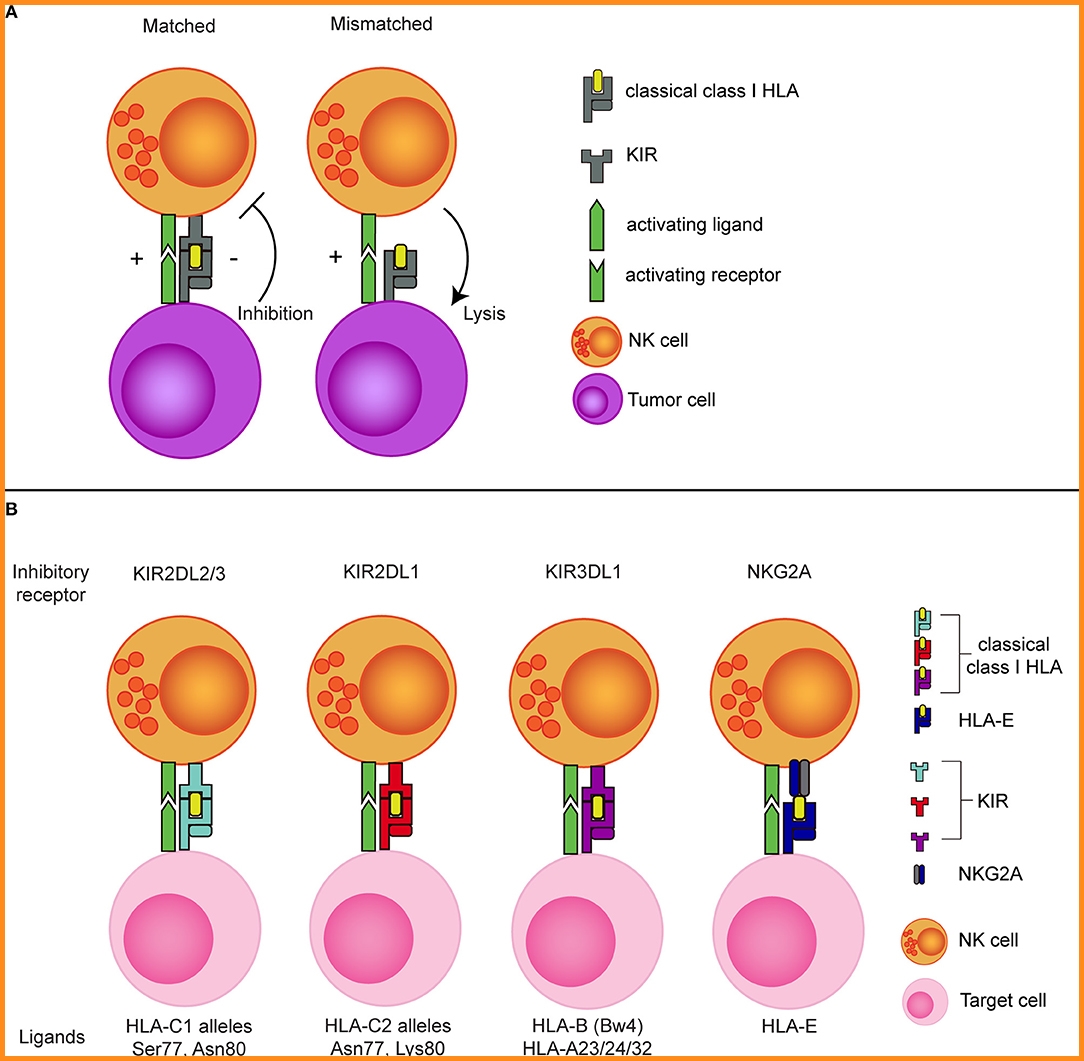
HLA class I molecules are the most important ligands for both the activating and the inhibitory KIRs. The best characterized inhibitory KIRs are: KIR2DL1, binding to HLA-C group 2 (C2) alleles having a lysine at position 80; KIR2DL2/3, interacting with HLA-C group 1 (C1) alleles having an asparagine at position 80. KIR3DL1, binding HLA-B alleles bearing a Bw4 motif as well as HLA-A*23/*24/*32. KIR3DL2 has been shown to interact with HLA-A*3/*11 and HLA-F. The activating KIR2DS1 and KIR2DS2 have been shown to bind with C2 and C1 alleles, respectively, and KIR2DS4 interacts with subsets of HLA-C alleles and with HLA-A*11. The ligands for the other KIRs remain elusive so far (Fig 5) (10).
Figure 5: The concept of NK cell alloreactivity concept based on interaction with HLA class I. (A) When an inhibitory KIR binds to a “matched” classical class I HLA molecule, an NK cell receives inhibitory signal from this interaction. In the absence of the corresponding class I HLA molecule (mismatched situation), the inhibitory signal is absent, resulting in a reduced NK cell activation threshold. (B) Inhibitory KIRs and NKG2A and their corresponding class I HLA molecules.
The potential of exploiting missing-self recognition to enhance the antitumor potential of NK cells in the allogeneic setting became most evident from the ground breaking work of Ruggeri et al. showing that a so called KIR ligand mismatch improved clinical outcome after haploidentical stem cell transplantation (haplo-SCT) in patients with acute myeloid leukemia (AML) (10,11).
For a long time, feasibility of haplo- or mismatched (MM) HSCT was limited by the high occurrence of post-transplant complications such as GVHD and infections. However, due to the recent successes of improved T cell depletion methods or by post-transplant administration of cyclophosphamide, haplo- and MM SCT became a feasible approach with a good safety profile and the major advantage that a large number of donors is usually available within the family or in registries (12).
In the haplo-SCT setting, patient and donor are matched based on one of the HLA haplotypes, meaning that half of the HLA genes are mismatched between patient and donor. This enables incompatibility between inhibitory KIRs, expressed on the donor NK cells, and their HLA ligands on patient tumor cells. As the donor KIR ligand mismatched NK cells in this setting will remain educated by the donor HLA background, they can efficiently detect missing-self and mediate more potent responses against the tumor cells than the non-mismatched NK cells that receive inhibitory signals via HLA (Fig 5) (10).
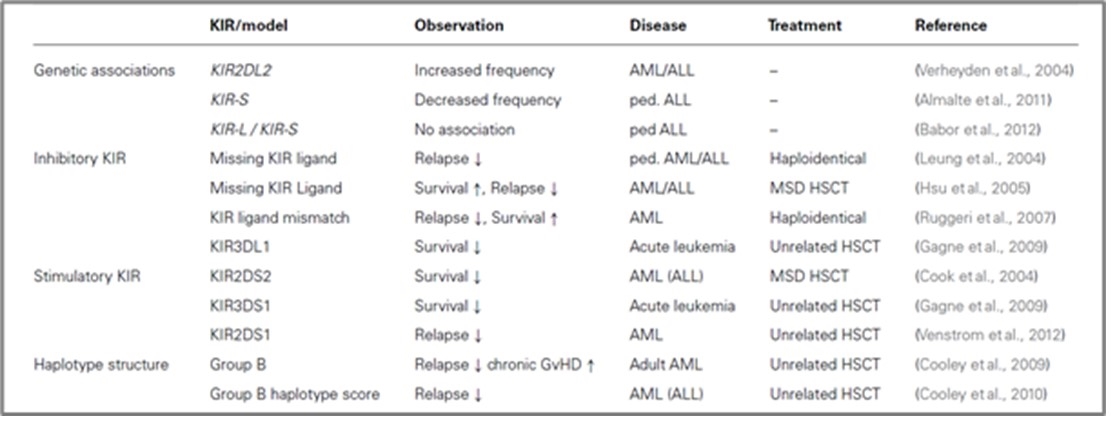
Although some subsequent studies did confirm the KIR ligand mismatch model, others could not find a beneficial effect of KIR ligand mismatching, which might be explained by specific differences in SCT protocols performed in the different transplant centres. Consequently, several modified donor selection models were suggested based on the consideration that donor NK cells expressing a certain inhibitory KIR would be alloreactive if the respective ligands (C1, C2, or Bw4) in the patient are missing. This model was originally proposed by Leung et al. (2004) and demonstrated better clinical outcome in haploidentical HSCT for pediatric lymphoid leukemia thereby proving that NK cell-mediated relapse control is not restricted to myeloid leukemia. Hsu et al. (2005) applied the same model to HLA-identical HSCT showing that patients with missing KIR ligands for which the donor possessed a specific KIR exhibited increased survival through decreased relapse incidence in AML and MDS patients. An overview of several studies investigating the role of KIR genes in SCT is given in Tab 1 (4,13,14).
Table 1. An overview of studies analyzing the role of KIR for susceptibility and clinical SCT for leukemia.
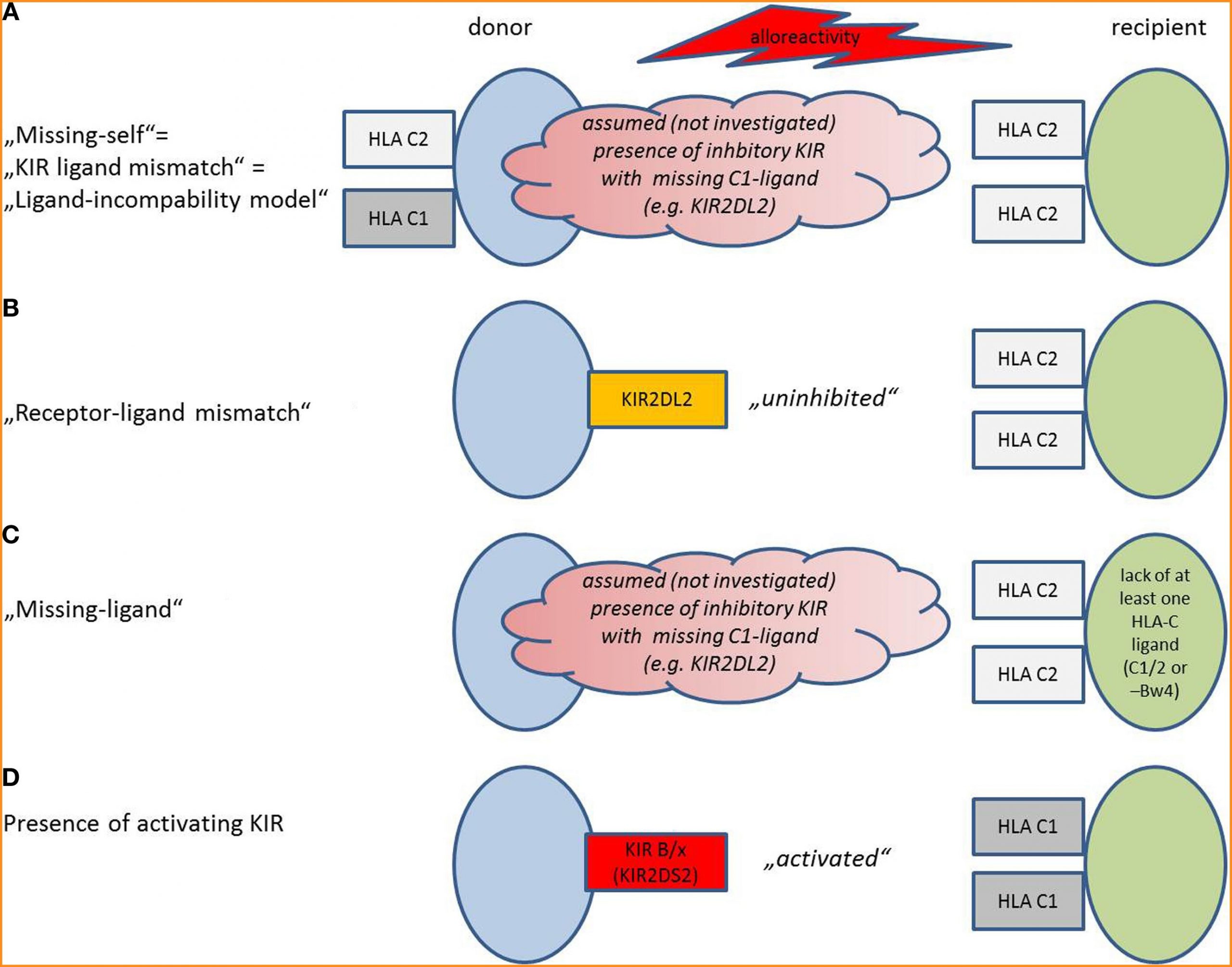
In general (also in the aforementioned studies), different definitions of a mismatch between the donor’s NK cells and the recipient’s HLA exist, depending on the method that was chosen for KIR and HLA (HLA-C1, C2, and Bw4) evaluation (Fig 6) (15).
Missing-Self/KIR-Ligand Mismatch (Figure 6A)
Alloreactivity was initially thought to be only dependent on lack of inhibitory HLA Class I molecules in the recipient that are present in the donor (“missing-self” or “KIR-ligand mismatch” or “ligand-incompatibility model”). For evaluation of KIR-ligand mismatch, donor and recipient are screened for expression of HLA: NK cells from a HLA C1/C1 donor will be alloreactive against a C2/C2 recipient. If a recipient expresses HLA-C1, C2, and Bw4, he will be resistant toward NK cell killing, as seen in one-third of the population. The presence of the respective inhibitory KIR in the donor is assumed but not verified.
Receptor–Ligand Mismatch (Figure 6B)
The receptor–ligand model states that donor NK cells become activated in the graft-versus-host direction; if they have inhibitory KIR, for which the HLA ligands in the recipient are missing, the NK cells become “uninhibited”. Thus, in addition to the HLA status of the recipient, confirmatory KIR genotyping of the patient is required. Other than in the first model, KIR on donor cells and HLA on recipient cells are investigated, not “assumed.” This model can be considered as an improvement of the “missing ligand model.”
Missing Ligand Model (Figure 6C)
Here, only the recipients’ HLA are genotyped, and missing HLA-C1, C2, or Bw4 for inhibitory KIR predict an alloreactivity of the graft; the presence of the respective KIR that would bind to the missing HLA is only assumed.
Presence of Activating KIR (Figure 6D)
Some theories emphasize that to achieve NK cell alloreactivity, “uninhibition” of NK cells by missing inhibitory HLA ligands might not be sufficient. Activation requires additional stimulation of activating KIR in the graft. In this model, alloreactivity can be predicted by measurement of activating KIR on donor cells. KIR haplotype B/x contains more activating KIR than KIR haplotype A/A. Some studies increase the predictive validity by detection of the respective activating ligands on donor cells.
Figure 6: Model situations that provoke NK cell alloreactivity.
To be eligible for KIR ligand mismatched transplantation, patients should genotypically lack expression of at least one of the inhibitory KIR ligands, meaning that they should miss either HLA-C1, -C2, or -Bw4 or a combination. However, 1/3 of the population express all three ligands. In addition to the absence of C1, C2, or Bw4 in the patient, the selected donor should express the HLA ligand that is missing in the patient to make sure the NK cells will be educated to sense the missing ligand and the corresponding KIR should be present on the cell surface. Especially for KIR3DL1 this is important to confirm, preferably by flow cytometry, as null alleles frequently occur.
KIR genes have two unusual features that could be harnessed for improving current HSCT strategies: firstly, KIR genes are highly polymorphic leading to novel opportunities beyond classical HLA matching strategies for selection of optimal stem cell donors based on KIR genetic diversity. Secondly, KIRs are expressed in a clonally distributed manner, which leads to the formation of NK cell repertoires encompassing tolerant as well as potentially alloreactive NK cell clones. Both features have a great potential to be exploited in the setting of clinical stem cell transplantation.
However, for multiple different diseases, currently there is still no real consensus on whether or not a KIR ligand mismatch has a clinical benefit and different output of studies shows that presumably this is highly dependent on the exact conditioning and transplantation protocol. Progress in research related to bone and peripheral blood stem cell transplantation, especially with the increasing reports of success rates in haploidentical transplantations for various clinical disorders, KIR genotype profiling may soon find its place in histocompatibility laboratory services.
References:
- Middleton D, Williams F, Halfpenny IA. KIR genes. Transpl Immunol 2005; 14:135–42.
- Miller, J. S., Cooley, S., Parham, P., Farag, S. S.,Verneris, M. R.,Mcqueen, K. L., et al. (2007). Missing KIR ligands are associated with less relapse and increased graft-versus-host disease (GVHD) following unrelated donor allogeneic HCT. Blood 109, 5058–5061.
- Barrow AD, Trowsdale J. The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunological Reviews (2008) 224:98-123
- Babor F, Fischer JC, Uhrberg M. The role of KIR genes and ligands in leukemia surveillance. Frontiers in immunology 2013; 4:27
- Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev (2002) 190:40-52
- Vilches, C., and P. Parham. 2002. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu. Rev. Immunol. 20:217–251.
- https://www.ebi.ac.uk/ipd/kir/alleles.html
- Jiang W, Johnson C, Jayaraman J, Simecek N, Noble J,Moffatt MF, et al. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. (2012)22:1845–54.
- Pende D, Falco M, Vitale M, Cantoni C, Vitale C, Munari E, et al. Killer Ig-Like receptors (KIRs): their role in NK cell modulation and developments leading to their clinical exploitation. Front Immunol. (2019) 10:1179
- Mahaweni NM, Ehlers FAI, Bos GMJ, Wieten L. Tuning natural killer cell anti-multiple myeloma reactivity by targeting inhibitory signaling via KIR and NKG2A. Front Immunol. 2018;9:2848.
- Ruggeri L, Mancusi A, Capanni M, Urbani E, Carotti A, Aloisi T, et al. Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood (2007) 110:433–40.
- Haas P, Loiseau P, Tamouza R, Cayuela JM, Moins-Teisserenc H, Busson M, et al. NK-cell education is shaped by donor HLA genotype after unrelated allogeneic hematopoietic stem cell transplantation. Blood (2011) 117:1021–9.
- Giebel, S., Locatelli, F., Lamparelli, T., Velardi, A., Davies, S., Frumento, G., et al. (2003). Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood 102, 814–819.
- Miller, J. S., Cooley, S., Parham, P., Farag, S. S.,Verneris, M. R.,Mcqueen, K. L., et al. (2007). Missing KIR ligands are associated with less relapse and increased graft-versus-host disease (GVHD) following unrelated donor allogeneic HCT. Blood 109, 5058–5061.
- Heidenreich S, Kröger N. Reduction of relapse after unrelated donor stem cell transplantation by KIR-based graft selection. Front Immunol. 2017;8:41.