Development of techniques in molecular biology over the last couple of years has not influenced only rEvolution (revolution + evolution) in HLA typing but also in other segments of onco-diagnostics and transplant medicine. Evaluation and implementation of Next Generation Sequencing (NGS) technology in Minimal Residual Disease (MRD) detection and monitoring have been one of the most active areas of research in this field.
MRD is defined as the tumor mass still remaining in the patient after chemotherapy or hematopoietic stem cell transplantation (HSCT). There is increasing evidence that early detection of subclinical levels of residual disease, or MRD, provides additional prognostic information. Thus, MRD follow-up is considered crucial for the identification of patients at high risk of relapse and has important clinical implications in both the pre- and post-HSCT setting in different types of blood cancer, such as acute myeloid leukemia (AML), chronic lymphocytic leukemia (CLL), multiple myeloma (MM) or myelodysplastic syndrome (MDS).
AML is known for its heterogeneity and aggressive malignancy of clonal, immature myeloid cells, thus this form of blood cancer has been a significant research focus of NGS applications in this field. The treatment of AML over the past four decades has remained mostly unchanged and the prognosis for the majority of patients has remained poor. It is believed that relapse in AML is in part driven by MRD that remains in the patient following treatment. Thus, there is a significant need for a sensitive and objective technology for MRD detection. Technologies such as multiparameter flow cytometry (MFC), quantitative real-time PCR (RQ-PCR), digital PCR (dPCR), or NGS have been employed to evaluate their utility in MRD assessment. (1)
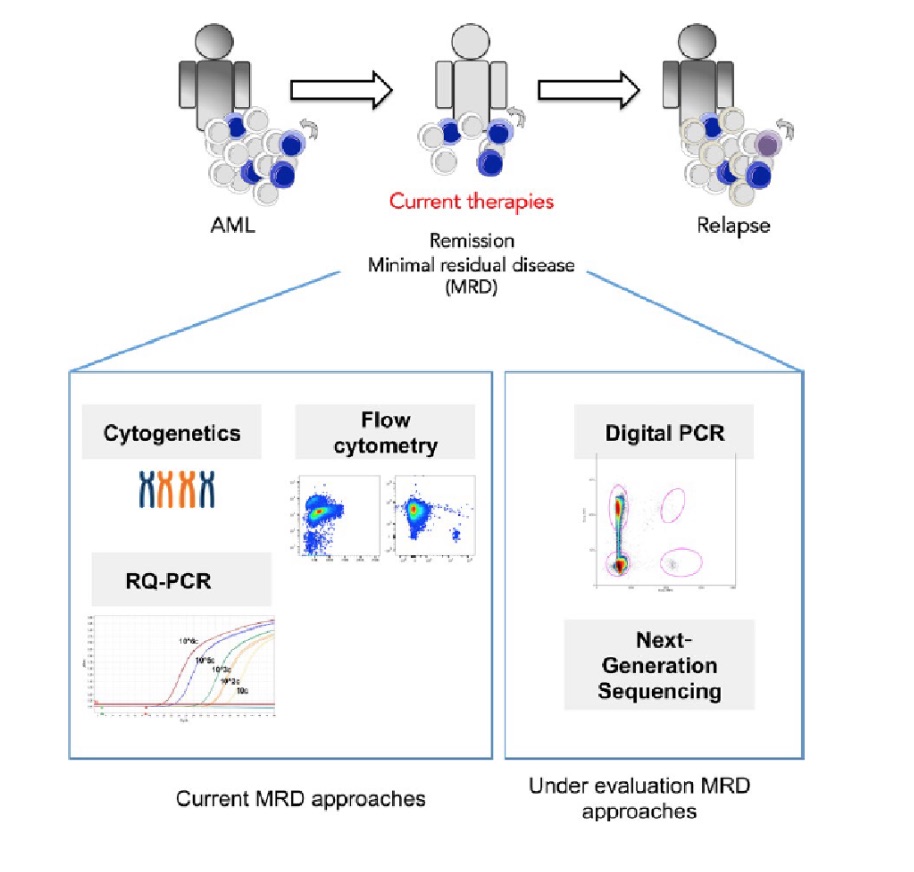
Figure 1. Current MRD assessment techniques include: cytogenetics, RQ-PCR, and flow cytometry. Newer techniques such as digital PCR and next generation sequencing are still under evaluation.1
Let’s do a quick overview of the aforementioned techniques and outline their pros and cons in MRD detection.
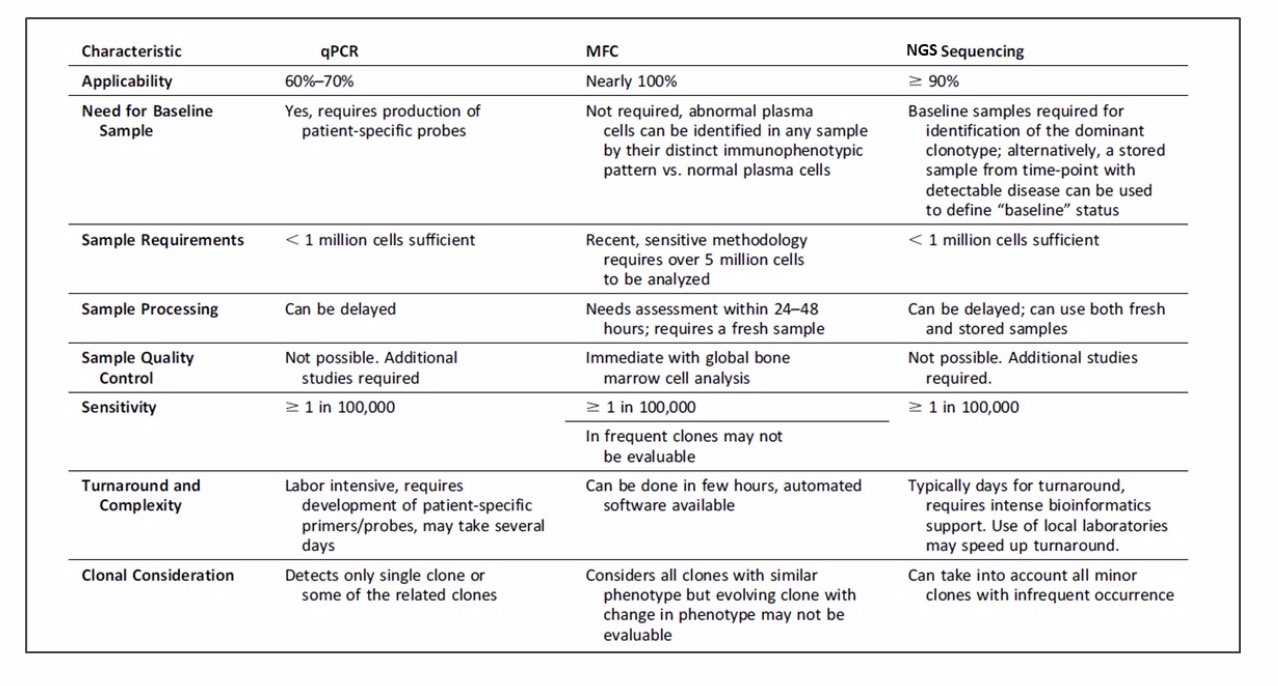
- REAL-TIME QUANTITATIVE PCR – This technique is able to identify chimeric fusion, gene rearrangements, genetic alterations, and differentially expressed genes. This diagnosis is useful and highly sensitive for two subgroups of patients – about 25% of the patients that exhibit chimeric fusion of the genes PML-RARA, AML1-ETO, or CBFB-MYH11 and about 45% of AML patients with cytogenetically normal karyotype that have mutations in NPM1, CEBPA, and FLT3-ITD. Being the gold standard in many labs, this method’s main drawbacks are sensitivity (depends on several factors, including the type of rearrangement, size of the junctional region and amount of DNA available for each reaction), feasibility and reproducibility. (2, 3)
- DIGITAL PCR (dPCR) – A single PCR is partitioned into hundreds to millions of droplets or wells (depending on the technological platform) each containing a single or few copies of the target template. The partitioned reaction undergoes thermocycling with each of these partitions constituting an individual reaction during the cycling process. Fluorescent signal is measured after an end-point PCR amplification for each partition individually. Thus, the number of positive molecules is determined from counting the number of successfully amplified fluorescent partitions rather than reaction kinetics as is the case with RQ-PCR. In addition, through absolute counting, the technique obviates the requirement for plasmid standards as is required for RQ-PCR. This technique has demonstrated promising results in the monitoring of MRD in hematological malignancies using both RNA-based and DNA-based methods. The sensitivity of dPCR is comparable to RQ-PCR and has demonstrated special promise for detecting single nucleotide variations (SNVs) due to the greater capability to differentiate the mutant versus normal allele in the absence of competing normal allele in each partition. (4, 5)
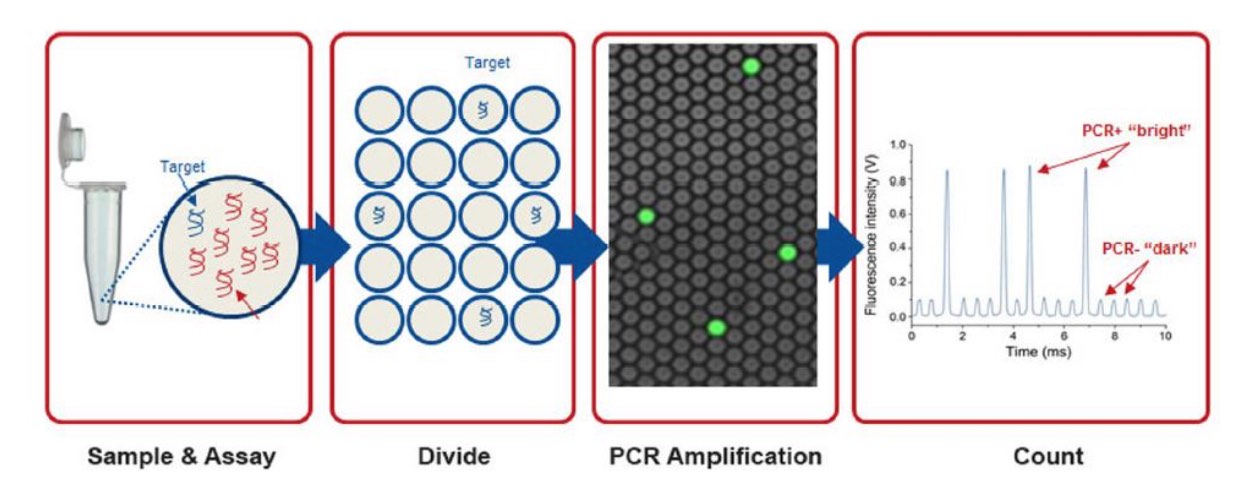
Figure 2. Separation and digital counting provide sensitive, absolute quantification. Digital PCR is performed by dividing the sample and the assay (e.g., qPCR hydrolysis probe and primers) into enough separate reaction chambers such that any reaction will contain either only 0 or 1 target molecule. Standard endpoint PCR is performed and the number of fluorescent reactions counted. PCR-positive, “bright” reactions each contained 1 target molecule, and PCR-negative, “dark” reactions have no target. 6
- MULTIPARAMETER FLOW CYTOMETRY (MFC) – This technique is based on antigen expression patterns that characterize the diverse lineages of normal hematopoietic cells. AML blasts have aberrant antigen expression patterns that are not detectable on the surface of bone marrow cells from healthy donors. These distinct immunophenotypic patterns are called leukemia-associated immunophenotypes and are present in 80% to 100% of AML patients. The prognostic significance of MRD detection using MFC has been examined in numerous studies performed in adults and children. (4,17,24,25,27–33) Overall, these studies demonstrate that MFC-detected MRD is prognostically significant in virtually all AML patients over a wide array of patient populations and treatment strategies. However, direct comparison of these studies is limited by variation in methodology, thresholds of positivity, patient population and treatment strategies. There have also been several contradictory findings regarding issues such as optimal timing of measurement (for example: post-induction vs post-consolidation) as well as cut-off values for MRD positivity. Despite this, several generalizations regarding the advantages and limitations of MRD measurement by MFC can be made. (9)
- NEXT GENERATION SEQUENCING – The use of single target assays for molecular MRD detection remains dominant in the field. However, given well-known patterns of clonal evolution evident in cancers and the need for highly personalized assays in patients with no recurrent or common lesions, there is a need for economical alternatives that enable more patients to be assessed for MRD in a manner that is robust to clonal evolution (molecular MRD) or phenotype switching (flow cytometry MRD). Going forward, next-generation sequencing (NGS) represents a potentially powerful alternative. Patients with no established molecular markers represent 42%, 38%, and 68% of patients with ages <15, 15-60, and >60 years old, respectively. Furthermore, due to the heterogeneity of the mutation repertoire in AML and the lack of hot spots in important but frequently mutated genes, it seems unfeasible to develop standardized assays on a per-patient basis. In addition, when tracking a single mutation, it has been observed that some AML patients will relapse with a different mutant clone, and in these cases, the marker being monitored is not informative to predict relapse. With this technique, there is no need for patient-specific assays as practically all mutations are detected. Recent studies report the use of NGS alone or in parallel with RQ-PCR for MRD detection. One caveat of NGS is that has limited its use for MRD assessment in the past few years, is the sequencing error rate and its impact on the sensitivity of the technique compared to the previously discussed methods. Recently, the introduction of error-corrected read technologies has helped overcome this limitation and greatly improving its sensitivity (lower than 0.03%). Another approach is the use of Bayesian analytical techniques for mutation calling informed by site-specific error rates and prior clinical data regarding mutation frequencies. However, even the advantages of error-corrected reads are likely to be eroded in certain genes by mappability limitations and errors arising from potential factors including gene paralogs. In addition to error-corrected reads, further improvements in read length and read mapping algorithms coupled with statistically principled variant calling techniques are likely to extend the sensitivity, specificity, and thus overall utility of NGS in MRD monitoring. Overall, NGS-based methods have the potential to detect subclinical disease in AML samples that do not qualify for the established leukemia-specific RQ-PCR or dPCR assays. In addition, NGS provides the ability to detect new emerging therapy-related leukemias that would otherwise be missed. Like MFC, NGS provides accurate information about leukemic tumor burden, an important parameter to measure treatment response. Validation and standardization in large clinical AML trials will be necessary. However, NGS opens the possibility to measure MRD in large patient cohorts. (1, 4, 10, 11)
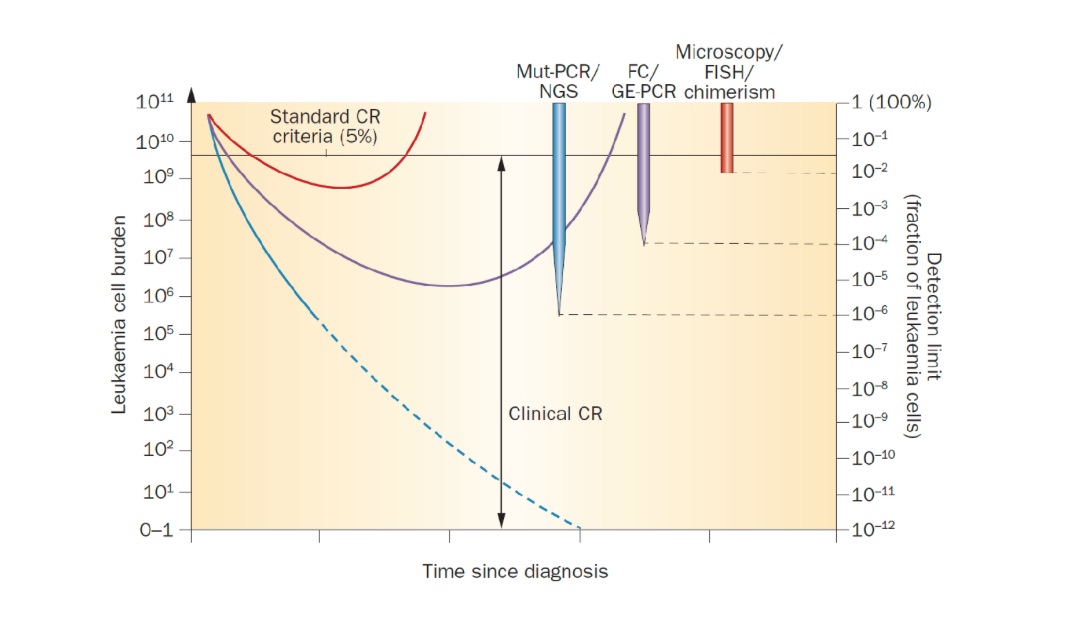
Figure 3. Detection thresholds of various MRD modalities compared to traditional clinical complete remission. Response criteria to assess the efficiency of treatment are limited by the intrinsic detection limit of the test being used. Conventional clinical CR status represents a wide range of potential disease burden. Microscopic interpretation of morphology by an expert pathologist can detect, at best, a two log reduction in disease burden (1% limit of detection), while FC and PCR for overexpressed genes have up to two log greater sensitivity (that is, ability to detect 1 abnormal cell in 10,000). PCR for fusion transcripts and specific mutations by PCR (Mut-PCR) or NGS can detect residual disease with approximately 10,000-fold greater sensitivity than traditional CR criteria. (12)
As described above, various techniques have been developed and utilized for detection of MRD. Each of them has advantages and disadvantages that must be taken into consideration. The ideal MRD test should fulfill several relevant characteristics:
- high applicability (useful among most patients)
- high sensitivity and specificity
- excellent feasibility (results can be obtained for most patients)
- easily accessible
- requirement for a sample to be transported with ease
- reproducibility
- proven clinical value
While none of these tests fully satisfy all of the ideal characteristics currently, NGS and improved MFC fulfill most of them and can be translated into an advanced platform that can be uniformly applied. Inclusion of both methodologies should be done in prospective trials to collect data that would allow the better understanding of the advantages and disadvantages of the individual approaches as well as the sensitivity of detection required in various clinical settings.
In the era of novel agents and combination therapies achieving very high conventional complete remission rates, the MRD status is beginning to play an important role in predicting superior outcome. MRD may serve as a biomarker to inform therapy and as a surrogate for overall survival. Specifically, achieving MRD-negative status may become a goal of future studies using induction, transplant, consolidation, and/or maintenance therapies. With available technologies and ease of MRD measurement, additional large prospective studies should be performed to define the clinical significance of MRD and its impact on patient outcome. In our quest toward personalizing therapy for patients, it may now be possible to both assess and monitor MRD using standardized assays and decide both intensity and length of therapy for individual patients to improve patient outcomes. (13)
The wealth of data recently generated highlights that MRD–negative status can be achieved in a large proportion of patients. Several studies clearly suggest significant improvement in both event-free survival and overall survival among those patients achieving MRD–negative status, especially with sensitivity of one cell in 1 million bone marrow cells. There is an evolving consensus that achieving MRD–negative status should become the ultimate goal of therapeutic intervention. Further future efforts should now be directed at determining how MRD status can be used to guide and personalize further therapy including type of consolidation and maintenance therapy.
References:
- N.M. Cruz, N. Mencia-Trinchant, D.C. Hassane, M.L. Guzman, Minimal residual disease in acute myelogenous leukemia, Int. J. Lab. Hematol. 39 (2017) 53–60
- Abdelhamid E, Preudhomme C, Helevaut N, et al. Minimal residual disease monitoring based on FLT3 internal tandem duplication in adult acute myeloid leukemia. Leuk Res. 2012;36:316‐323.
- Schlenk RF, Döhner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909‐1918.
- Grimwade D, Freeman SD. Defining minimal residual disease in acute myeloid leukemia: which platforms are ready for “prime time”? Blood. 2014;124:3345‐3355.
- Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003‐1005.
- IDT- Digital PCR (dPCR) – What is it and why use it?
- Kern W, Bacher U, Haferlach C, Schnittger S, Haferlach T. The role of multiparameter flow cytometry for disease monitoring in AML. Best Pract Res Clin Haematol. 2010;23:379‐390.
- Vidriales B, Coustan-smith E, Campana D, Orfao A, San-Miguel JF. Minimal residual disease monitoring by flow cytometry. Best Pract Res Clin Haematol. 2003;16:599‐612.
- Jaso JM, Wang SA, Jorgensen JL, Lin P. Multi-color flow cytometric immunophenotyping for detection of minimal residual disease in AML: past, present and future. Bone Marrow Transplant. 2014;49:1129‐1138.
- Gerstung M, Papaemmanuil E, Campbell PJ. Subclonal variant calling with multiple samples and prior knowledge. Bioinformatics. 2014;30:1198‐1204.
- Thol F, Kölking B, Damm F, et al. Next-generation sequencing for minimal residual disease monitoring in acute myeloid leukemia patients with FLT3-ITD or NPM1 mutations. Genes Chromosom Cancer. 2012;51:689‐695.
- Hourigan, C. S. & Karp, J. E. Nat. Rev. Clin. Oncol. 10, 460–471 (2013)
- Herve Avet-Loiseau Minimal Residual Disease by Next-Generation Sequencing: Pros and Cons American Society of Clinical Oncology Educational Book 2016 :36, e425-e430
Written by Vera Siffnerova (FAS, Omixon)