Introduction
The Illumina NGS sequencing fleet includes instruments of different sample capacity. One of them is the MiniSeq – designed for laboratories with a small to medium sample throughput. Omixon’s Holotype HLA was initially developed and validated for Illumina’s MiSeq, as this is a commonly available instrument in clinical laboratories. However, smaller HLA laboratories that aim to implement NGS in their routine, benefit from the smaller-sized and more economical MiniSeq.
MiniSeq Instrument
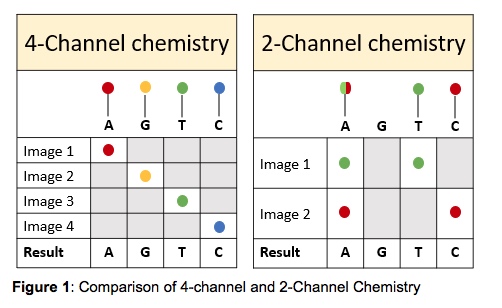
The MiniSeq uses a 2-channel SBS (sequencing by synthesis) chemistry which means that there are only two fluorescent dyes needed for the nucleotide detection, as opposed to the MiSeq that uses a 4-channel chemistry. The MiniSeq takes only two images per sequencing cycle allowing for easier signal detection and faster image analysis. The images are taken using red and green filter bands. Thymines are labeled with a green fluorophore, cytosines are labeled with a red fluorophore and adenines are labeled with both red and green fluorophores. Guanines are permanently dark (Figure 1). This makes the MiniSeq a more cost-effective device than other sequencing instruments.
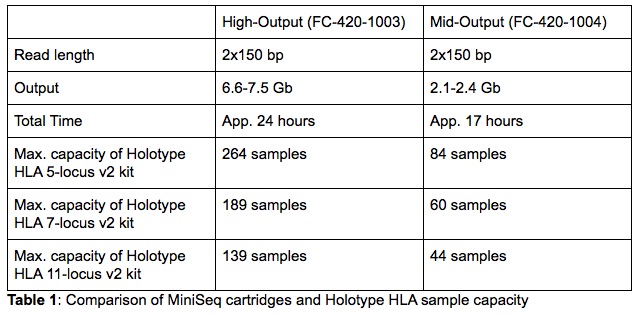
Additionally, the MiniSeq has a small laboratory footprint which makes it a very convenient solution for laboratories struggling with space. Despite the small dimensions, the MiniSeq’s capacity is sufficient to satisfy the needs of a large number of HLA laboratories processing both solid organ and bone marrow transplant samples. Just like the MiSeq, the MiniSeq supports a 300-cycle chemistry making it fully compatible with Holotype HLA. Depending on the number of samples that are processed in a single library, one can choose between a High- or Mid-Output MiniSeq cartridge (full details in Table 1).
Validation of Holotype HLA on the MiniSeq
Holotype HLA was validated on the MiniSeq using a cohort of 100 samples, composed of DNA samples from patients and donors, as well as HLA EPT samples (External Proficiency Testing). In all cases, the DNA was extracted using automated methods (InnuPure from Analytik Jena, iPrep from Thermo Scientific, Arrow from Nordiag). The input DNA concentration for the Long-Range PCR was between 20 – 30 ng/ul. All PCR reactions and library preparation incubations were carried out in the Biometra TAdvanced thermocyclers (Analytik Jena), and following the Holotype HLA v2 protocol, with no deviation. The library size selection was performed using the bead-based size selection method and the final library concentration was estimated using the Qubit.
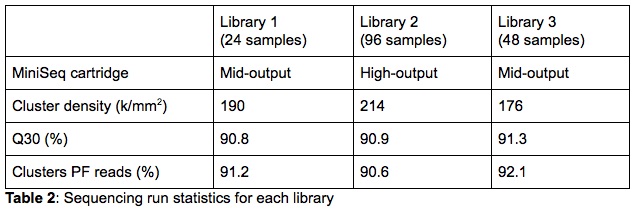
For the validation, both High- and Mid-Output 300-cycle sequencing cartridges were used. The purpose was to test libraries of different sizes to validate the capacity flexibility of the MiniSeq. Three sequencing libraries, each containing a different number of samples and loaded on a different type of cartridge, were run as part of the validation (Table 2). On the cartridge and capacity comparison, it is clear that the MiniSeq metrics are very comparable for all libraries. This indicates that neither the number of samples combined in a single library nor the cartridge size influence the sequencing data quality generated by the MiniSeq.
Most of the DNA samples had been previously typed by NGS and thus the majority of the reference typings were available at 3rd field resolution. However, the HLA EPT reference typings (a small subset of the tested cohort) were only available at 2nd field resolution. In order to avoid any bias in the comparison of the results, the 2nd field resolution was considered for all samples. Of the 100 samples that were tested as part of this validation, 62 were unique DNAs and 38 were run in duplicate to evaluate the inter-run and inter-technician variability.
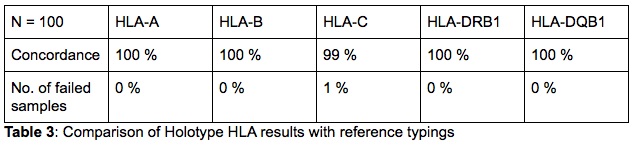
The results of all the samples and loci were found to be fully concordant with the reference typing on a 2nd field (Table 3). Only one discrepancy was found for a single locus. In one instance the reference typing for HLA-C of one sample was HLA-C*07:02, while the Holotype HLA result was HLA-C*07:18. Although there is no evidence or quality issues to indicate that the NGS result is incorrect, this locus was not re-tested with any method and thus has not been confirmed. Lastly, the samples that were run in duplicate, in different runs and by different technicians, all resulted in concordant typings with the reference typings.
All the above indicate that Holotype HLA when run on Illumina’s smaller sequencer, the MiniSeq, is well-suited for low to medium throughput HLA laboratories and can be easily adapted for various numbers of samples per run, depending on the laboratory’s needs.
Acknowledgements
We would like to specifically thank dr. Monica Dutescu from the National HLA Laboratory – Bucharest, Romania for the collaboration on this project and for providing us the data.
Article written by Dr. Libor Kolesár (Senior Field Application Scientist, Omixon)