I’ve already written some posts about fasta files and reference sequences (about file formats and where to get reference sequences.
You might remember from my first “Workflow Wednesdays” post, that I selected short reads created with the three major sequencing technologies from the same E. coli strain: strain K-12 substrain MG1655.
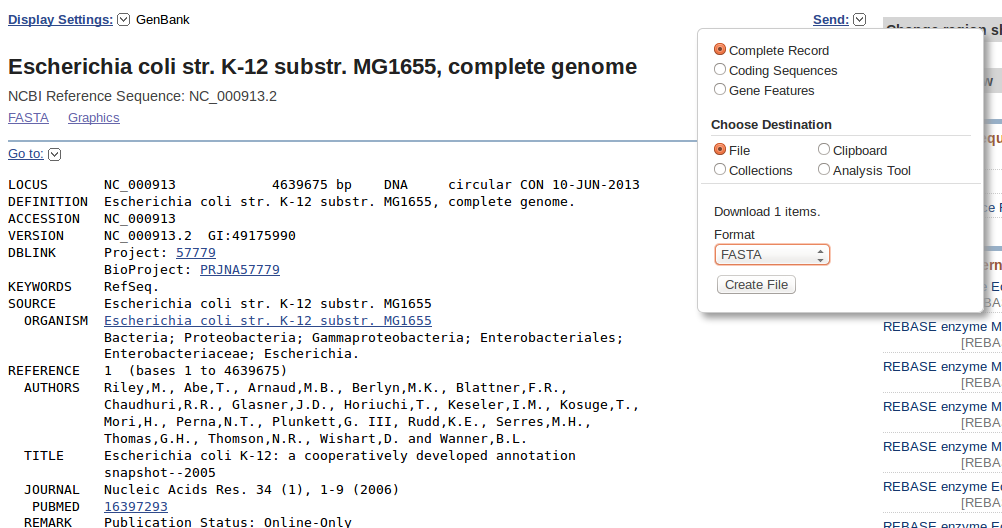
The easiest way to get the genome sequence of this “bug”, is possibly the NCBI nucleotide archive (or one if its sister sites). So let’s go to NCBI, select the Nucleotide database and search. With a little searching around, you should find this entry. You can download the sequence or the whole record in different formats with the “Send” function. You’ll need the reference in fasta format, so select that one.
The file is automatically named as “sequence.fasta”, move it to your work folder and rename it:
mv /home/rk/Downloads/sequence.fasta /home/rk/tasks/blog/workflow/NC_000913.2.fa
It’s usually a good idea, to name your sequence file after the accession number, so you don’t end up with several copies of the same sequence.
You can always look at the header of the fasta which – at least for files downloaded from NCBI – contains some additional information. So let’s take a look at the beginning of the file!
$head NC_000913.2.fa >gi|49175990|ref|NC_000913.2| Escherichia coli str. K-12 substr. MG1655, complete genome AGCTTTTCATTCTGACTGCAACGGGCAATATGTCTCTGTGTGGATTAAAAAAAGAGTGTCTGATAGCAGC TTCTGAACTGGTTACCTGCCGTGAGTAAATTAAAATTTTATTGACTTAGGTCACTAAATACTTTAACCAA TATAGGCATAGCGCACAGACAGATAAAAATTACAGAGTACACAACATCCATGAAACGCATTAGCACCACC ATTACCACCACCATCACCATTACCACAGGTAACGGTGCGGGCTGACGCGTACAGGAAACACAGAAAAAAG CCCGCACCTGACAGTGCGGGCTTTTTTTTTCGACCAAAGGTAACGAGGTAACAACCATGCGAGTGTTGAA GTTCGGCGGTACATCAGTGGCAAATGCAGAACGTTTTCTGCGTGTTGCCGATATTCTGGAAAGCAATGCC AGGCAGGGGCAGGTGGCCACCGTCCTCTCTGCCCCCGCCAAAATCACCAACCACCTGGTGGCGATGATTG AAAAAACCATTAGCGGCCAGGATGCTTTACCCAATATCAGCGATGCCGAACGTATTTTTGCCGAACTTTT GACGGGACTCGCCGCCGCCCAGCCGGGGTTCCCGCTGGCGCAATTGAAAACTTTCGTCGATCAGGAATTT
By default, the “head” command prints the first 10 line of a file (and “tail” prints the last 10).
As you can see, this fasta file is nicely formatted with uniform line length and an informative header. If you have a fasta file which has non-uniform line length, you can easily fix it with Picard’s NormalizeFasta tool.
Picard also has another useful tool: you can create a sequence dictionary, with the CreateSequenceDictionary tool.
$java -jar /homerk//picard-tools-1.86/CreateSequenceDictionary.jar R=NC_000913.2.fa O=NC_000913.2.fa.dict [Fri Jun 28 12:32:25 CEST 2013] net.sf.picard.sam.CreateSequenceDictionary REFERENCE=NC_000913.2.fa OUTPUT=NC_000913.2.fa.dict TRUNCATE_NAMES_AT_WHITESPACE=true NUM_SEQUENCES=2147483647 VERBOSITY=INFO QUIET=false VALIDATION_STRINGENCY=STRICT COMPRESSION_LEVEL=5 MAX_RECORDS_IN_RAM=500000 CREATE_INDEX=false CREATE_MD5_FILE=false [Fri Jun 28 12:32:25 CEST 2013] Executing as rk@enceladus on Linux 3.8.0-25-generic amd64; Java HotSpot(TM) 64-Bit Server VM 1.6.0_38-b05; Picard version: 1.86(1363) [Fri Jun 28 12:32:25 CEST 2013] net.sf.picard.sam.CreateSequenceDictionary done. Elapsed time: 0.00 minutes. Runtime.totalMemory()=125173760
If you peek into the dict file, you can see, that it basically contains the reference related parts of a sam file header:
$less NC_000913.2.fa.dict @HD VN:1.4 SO:unsorted @SQ SN:gi|49175990|ref|NC_000913.2| LN:4639675 UR:file:/home/rk/tasks/blog/workflow/NC_000913.2.fa M5:05dc7a37701cdc6bcf154344a227983d
You can also generate a set of index files with BWA:
$bwa index NC_000913.2.fa [bwa_index] Pack FASTA... 0.06 sec [bwa_index] Construct BWT for the packed sequence... [bwa_index] 2.03 seconds elapse. [bwa_index] Update BWT... 0.02 sec [bwa_index] Pack forward-only FASTA... 0.05 sec [bwa_index] Construct SA from BWT and Occ... 0.70 sec [main] Version: 0.6.2-r126 [main] CMD: bwa index NC_000913.2.fa [main] Real time: 2.900 sec; CPU: 2.872 sec
SAMTools also have a fasta indexing function, which creates a .fai file:
samtools faidx NC_000913.2.fa
Some of these different files contain very basic information: the names and lengths of reference contigs in the fasta file. Others are somewhat more complicated (e.g. the FM-index created (and required) by BWA). But don’t worry, alignment and variant call tools usually “tell” you what kind of index file they require.