Short reads can be stored in several different formats. The best known (and most used) of these is the fastq format, which contains both the base and quality values for each read within a single file. Note, that there are different fastq formats around. The main difference between these is the calculation and representation of quality values.
Quality calculation
The “Sanger” fastq format uses the standard Phred-formula for quality calculation: QPHRED=-10xlog10(Pe), where Pe is the estimated probability of error (in this case, the estimated probability of the base the call being wrong). In the older versions (prior to version 1.3, to be exact) of Solexa/Illumina fastq files, a different quality calibration method was used:
According to the latest CASAVA documentation (v1.8.2), Illumina now uses the Sanger calculation method:
“A quality score (or Q-score) is a prediction of the probability of an incorrect base call. Based on the Phred scale, the Q-score serves as a compact way to communicate very small error probabilities. Given a base call, X, the probability that X is not true, P(~X), is expressed by a quality score, Q(X), according to the relationship:
Q(X) = -10 log10(P(~X)) where P(~X) is the estimated probability of the base call being wrong.”
You can find a very useful article, that describes different variations of the fastq format here.
Quality encoding
Quality values are converted to a single character using the ASCII table. As this table starts with 32 non-printing characters, the quality values are represented by the character equivalent to Q+A, where A is at least 33 the second half of the ASCII table. Different fastq versions used different A values (for details see below, source).
SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS.....................................................
..........................XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX......................
...............................IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII......................
.................................JJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJ......................
LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL....................................................
!"#$%&'()*+,-./0123456789:;<=>?@ABCDEFGHIJKLMNOPQRSTUVWXYZ[\]^_`abcdefghijklmnopqrstuvwxyz{|}~
| | | | | |
33 59 64 73 104 126
0........................26...31.......40
-5....0........9.............................40
0........9.............................40
3.....9.............................40
0........................26...31........41
S - Sanger Phred+33, raw reads typically (0, 40)
X - Solexa Solexa+64, raw reads typically (-5, 40)
I - Illumina 1.3+ Phred+64, raw reads typically (0, 40)
J - Illumina 1.5+ Phred+64, raw reads typically (3, 40)
with 0=unused, 1=unused, 2=Read Segment Quality Control Indicator (bold)
(Note: See discussion above).
L - Illumina 1.8+ Phred+33, raw reads typically (0, 41)
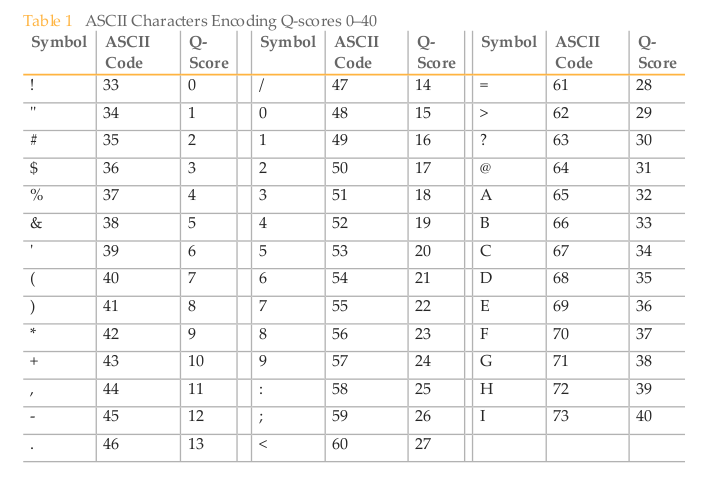
A different representation of the Illumina 1.8+ fastq quality encoding format from the CASAVA (v.1.8.2) User Guide.
Another file format for sequencing reads is SFF (Standard Flowgram Format), which is a binary format that contains trace information and used for both 454 and Ion Torrent reads. You can find detailed information about this file format here. Also, there’s an excellent post about 454 sff files on the Newbler blog. Some documentation about the Ion Torrent version is available on the Ion Community site (viewing this site requires a free registration).
Tip: Although SAM and BAM formatted files are primarily for storing reference based alignments, unaligned reads can be stored in these formats as well.






