Authors: Libor Kolesar, Matthias Niemann, Efi Melista
INTRODUCTION
In the previous blog post we talked about a hot topic in the HLA community right now which is the one of epitope-based matching. We discussed one of the methodologies that are commonly used, HLA Matchmaker, and this time we will be discussing a different methodology called PIRCHE. PIRCHE stands for Predicted Indirectly ReCognizable HLA Epitopes. While HLA Matchmaker is considering epitopes in the context of sites for direct recognition by B cells, PIRCHE is assessing epitopes that are targets for indirect recognition by T cells. In other words, the PIRCHE algorithm is a tool that has been developed for the identification of permissible mismatches in transplantation based on the prediction of epitope-mismatches that are present in HLA mismatched patient-donor pairs and that are involved in T-cell-mediated alloimmune responses.
But let’s first start with a little bit of the background on indirect recognition by T cells which plays an important role in alloreactivity.
INDIRECT T-CELL ALLORECOGNITION
T cells are known key players in both T-cell and B-cell mediated immune reactions. We can identify three pathways of T-cell interaction with allogeneic HLA molecules. 1) Direct recognition which means that T cells detect complete allogeneic HLA molecules on the surface of allogeneic cells. 2) Indirect recognition means that T cells detect epitopes of allogeneic HLA molecules processed and presented on self-HLA molecules located on self-antigen-presenting cells. 3) And semi-direct recognition in which case the T cells detect complete allogeneic peptide:HLA complexes exchanged between allogeneic and self-antigen-presenting cells (Fig. 1). The indirect recognition of mismatched HLA epitopes is very important for B cell mediated antibody production. B cells can internalize and process the mismatched HLA molecule and then display its fragments in the context of self-HLA molecule on their surface. The T helper cells then recognize these unknown peptides and start providing costimulatory signals to B cells by producing cytokines resulting in immunoglobulin isotypes switching from IgM to IgG, and maturing naive B cells into the mature plasma cells producing detrimental IgG donor specific HLA antibodies. These antibodies are the cause of antibody-mediated graft rejection.
Several studies have shown the key function of T-cell recognition both in the development of GVHD in HSC transplant settings or allograft rejection after organ transplantation (Benichou et al., 1992; Lee et al., 1994; Vella et al., 1997; Baker et al., 2001).
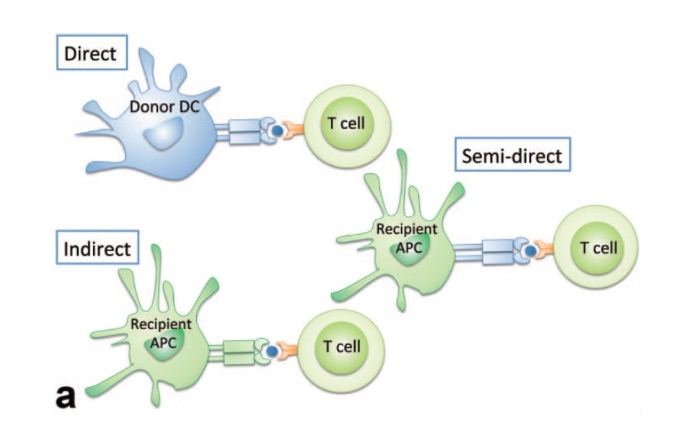
Figure 1: Three pathways of T-cell-mediated allorecognition. Adopted from https://wiki.mcmaster.ca/LIFESCI_4M03/group_1_presentation_1_-_kidney_transplant_rejection
PIRCHE ALGORITHM
Many studies have also shown that donor-recipient pairs that are not fully matched at HLA antigens may result in a successful transplant outcome (Kanda J., 2013; Heidth and Class, 2018). This was a very interesting observation and even though the background was not fully understood, it prompted investigations and the development of new methodologies for defining HLA compatibility. These studies have helped in the identification of permissible/acceptable and nonpermissible/unacceptable mismatches. Due to the high polymorphic nature of HLA, it is quite challenging to test each HLA mismatch in vitro or in vivo, thus researchers are looking for in-silico models for estimating the risk of HLA mismatches.
The PIRCHE algorithm has been developed for the prediction of indirect T-cell recognition. This algorithm predicts the number of peptides derived from every donor-recipient HLA mismatch that are then presented in the context of HLA class I or HLA class II molecules. Mismatched HLA-derived epitopes that can be presented on HLA class I molecules are designated as PIRCHE-I, whereas mismatched HLA-derived epitopes that can be presented on HLA class II molecules are designated as PIRCHE-II (Geneugelijk et al., 2014). Characterization of HLA mismatch permissibility prior a transplantation may assist in the selection of the most optimal recipient-donor pair and thus significantly decrease the risk of post-transplant immunological complications. Additionally, it can help in the donor search for re-transplantation giving the patient a higher chance of long-term survival.
The PIRCHE score can be calculated for every donor-recipient pair. This score reflects the level of indirect immune alloreactivity. CD8+ T cells recognize allopeptides presented on HLA class I antigens so PIRCHE-I score theoretically describes the level of CD8+ T cell alloreactivity. On the other hand, CD4+ T helper cells recognize allopeptides bound to HLA class II antigens. Thus, the PIRCHE-II score theoretically describes the level of CD4+ T cell alloreactivity.
To give you a better idea on how the PIRCHE algorithm works, let’s have a look at Figure 2. It represents an example of two cases of donor-recipient pairs and their PIRCHE-II score derived from different HLA molecules. Although both donor-recipient pairs have the same number of HLA mismatches at allele level, when considering the number of different HLA-derived peptides the second pair reaches a higher PIRCHE score which indicates a higher risk of donor-specific antibody formation.
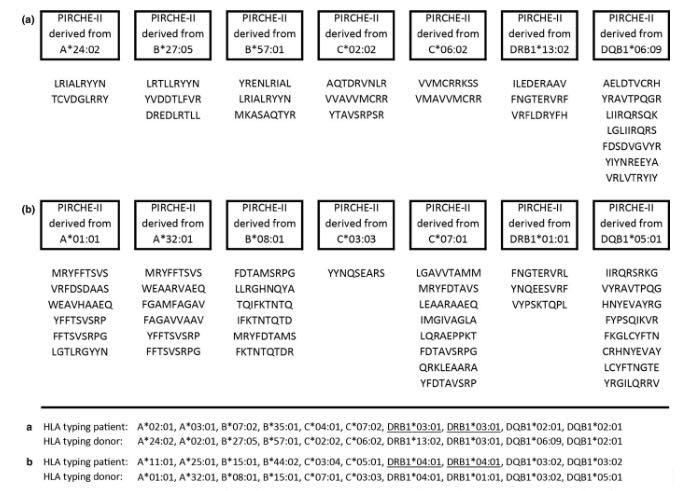
Figure 2: Two illustrative examples of PIRCHE-II core-peptides derived from different HLA molecules for different donor–recipient combinations. Adopted from Geneugelijk and Spierings, 2018.
The PIRCHE scoring system can be applied both to hematopoietic stem cell transplantation and solid organ transplantation. Lachmann et al. (2017) showed a HLA- and eplet-matching independent correlation between PIRCHE-II and dnDSA post kidney transplantation. Furthermore, Geneugelijk et al. (2018) confirmed, an increased PIRCHE score also impairs graft survival. Zheng et al. (2019) analyzed the distribution frequency of HLA antigen genes and antibodies and explored the relationship of the PIRCHE score with the DSA production and AMR (antibody-mediated rejection) occurrence in kidney transplant settings. They have shown that the PIRCHE score of the DSA+ and DSA+ / AMR+ groups were markedly higher than that of DSA- and DSA+ / AMR- groups respectively (P<0.05).
Daniëls at al (2018) carried out an elegant multicentre study whose aim was to perform an independent confirmation of earlier observations that HLA Matchmaker and PIRCHE II scores may be a better predictor for dnDSA formation than classical HLA antigen matching in kidney transplantations. They studied a cohort of retransplanted patients (2nd transplant only) whose graft failed due to the HLA mismatches from the 1st transplantation for better characterization and effect of dnDSA. They clearly showed that the dnDSA formation correlates with the number of HLA mismatches, HLA Matchmaker and PIRCHE scores, with high sensitivity and specificity which increased even more when other loci like HLA-C and -DQ were added. PIRCHE-II scores and HLA Matchmaker scores were the best predictors of dnDSA.
In hematopoietic transplants, Thus et al. (2014) have shown that PIRCHE-I or -II derived from DPB1 mismatches increase the risk of acute GvHD compared to patients with no PIRCHE-I/-II. Recently, Geneugelijk et al. (2019) showed that the PIRCHE approach could also be applied to other mismatching loci. Stem cell transplanted patients with an early disease stage showed significantly impaired overall survival when the patient-derived PIRCHE-II score was high. However, patients with low PIRCHE-II scores had similar survival rates as patients transplanted with a 10/10 HLA matched donor.
In the field of haploidentical stem cell transplantation, there is no evidence for an effect by PIRCHE matching yet. The study of Huo et al. (2018) was investigating whether indirect recognition of mismatched HLA antigens could predict the clinical outcomes in haploidentical stem cell transplantation but no significant associations were observed between PIRCHE-Ⅰ or PIRCHE-Ⅱ scores and the clinical outcomes. It remains unclear, whether effects were masked by confounder effects or whether the treatment protocol levels out a potential effect by PIRCHE.
CONCLUSION
With the last two blog posts of our series, we aimed to show the different approaches of HLA matching and how matching based on the functional aspects of HLA can be utilized and result in successful transplantation outcomes. To date, there is significant evidence of the superiority or epitope matching as opposed to HLA matching and both PIRCHE and HLA Matchmaker have proved to be useful in-silico tools for assessing the risk of immunological complications after transplantation. We are expecting more and more new studies evaluating PIRCHE as a new tool, which will show whether it can be world-widely implemented into the allocation schemes of transplant centers.
We will certainly be keeping up with the HLA community and literature to see how epitope matching will impact the transplantation decision making. Until then, stay tuned for another interesting topic next month!
REFERENCES
Baker RJ, Hernandez-Fuentes MP, Brookes PA, Chaudhry AN, Cook HT, Lechler RI. Loss of direct and maintenance of indirect alloresponses in renal allograft recipients: implications for the pathogenesis of chronic allograft nephropathy. J Immunol. 2001 Dec 15;167(12):7199-206.
Benichou G, Takizawa PA, Olson CA, McMillan M, Sercarz EE. Donor major histocompatibility complex (MHC) peptides are presented by recipient MHC molecules during graft rejection. J Exp Med. 1992 Jan 1;175(1):305-8.
Daniëls L, Naesens M, Bosmans JL, Abramowicz D, Nagler E, Van Laecke S, Peeters P, Kuypers D, Emonds MP. The clinical significance of epitope mismatch load in kidney transplantation: A multicentre study. Transpl Immunol. 2018 Oct;50:55-59
Geneugelijk K, Thus KA, Spierings E. Predicting alloreactivity in transplantation. J Immunol Res. 2014.
Geneugelijk K, Spierings E. Matching donor and recipient based on predicted indirectly recognizable human leukocyte antigen epitopes. Int J Immunogenet. 2018 Apr;45(2):41-53.
Geneugelijk K, Niemann M, Drylewicz J, van Zuilen AD, Joosten I, Allebes WA, van der Meer A, Hilbrands LB, Baas MC, Hack CE, van Reekum FE, Verhaar MC, Kamburova EG, Bots ML, Seelen MAJ, Sanders JS, Hepkema BG, Lambeck AJ, Bungener LB, Roozendaal C, Tilanus MGJ, Vanderlocht J, Voorter CE, Wieten L, van Duijnhoven EM, Gelens M, Christiaans MHL, van Ittersum FJ, Nurmohamed A, Lardy JNM, Swelsen W, van der Pant KA, van der Weerd NC, Ten Berge IJM, Bemelman FJ, Hoitsma A, van der Boog PJM, de Fijter JW, Betjes MGH, Heidt S,
Geneugelijk K, Thus KA, van Deutekom HWM, Calis JJA, Borst E, Keşmir C, Oudshoorn M, van der Holt B, Meijer E, Zeerleder S, de Groot MR, von dem Borne PA, Schaap N, Cornelissen J, Kuball J and Spierings E (2019) Exploratory Study of Predicted Indirectly ReCognizable HLA Epitopes in Mismatched Hematopoietic Cell Transplantations. Front. Immunol. 10:880. doi: 10.3389/fimmu.2019.00880
Roelen DL, Claas FH, Otten HG, Spierings E. PIRCHE-II Is Related to Graft Failure after Kidney Transplantation. Front Immunol. 2018 Mar 5;9:321.
Heidt S, Claas FHJ. Transplantation in highly sensitized patients: challenges and recommendations. Expert Rev Clin Immunol. 2018 Aug;14(8):673-679.
Huo MR, Li D, Chang YJ, Xu LP, Zhang XH, Liu KY, Huang XJ. Predicted indirectly recognizable HLA epitopes are not associated with clinical outcomes after haploidentical hematopoietic stem cell transplantation. Hum Immunol. 2018 Feb;79(2):117-121.
Kanda J. Effect of HLA mismatch on acute graft-versus-host disease. Int J Hematol. 2013 Sep;98(3):300-8.
Lachmann N, Niemann M, Reinke P, Budde K, Schmidt D, Halleck F, Pruß A, Schönemann C, Spierings E, Staeck O. Donor-Recipient Matching Based on Predicted Indirectly Recognizable HLA Epitopes Independently Predicts the Incidence of De Novo Donor-Specific HLA Antibodies Following Renal Transplantation. Am J Transplant. 2017 Dec;17(12):3076-3086.
Lee RS, Grusby MJ, Glimcher LH, Winn HJ, Auchincloss H Jr. Indirect recognition by helper cells can induce donor-specific cytotoxic T lymphocytes in vivo. J Exp Med. 1994 Mar 1;179(3):865-72.
Thus KA, Ruizendaal MT, de Hoop TA, Borst E, van Deutekom HW, Te Boome L, Kuball J, Spierings E. Refinement of the definition of permissible HLA-DPB1 mismatches with predicted indirectly recognizable HLA-DPB1 epitopes. Biol Blood Marrow Transplant. 2014 Nov;20(11):1705-10.
Vella JP, Spadafora-Ferreira M, Murphy B, Alexander SI, Harmon W, Carpenter CB, Sayegh MH. Indirect allorecognition of major histocompatibility complex allopeptides in human renal transplant recipients with chronic graft dysfunction. Transplantation. 1997 Sep 27;64(6):795-800.
Zheng J, Kuang PD, Zhang Y, Zhao Q, He XL, Ding XM, Xue WJ. [Relationship of distribution frequency of HLA antigen/antibody and PIRCHE score with DSA production and AMR occurrence]. Zhonghua Yi Xue Za Zhi. 2019 Mar 26;99(12):901-906.






