Written by Dr. Libor Kolesár (Senior Field Applications Scientist, Omixon)
Introduction
Most transplant laboratories’ protocols for donor/patient matching are based on the number of mismatches between patient and donor HLA alleles/antigens. The fewer mismatches between two individuals, the better. In solid organ transplantation (SOT) minimum matching refers to matching on antigen level, while in stem cell transplantation (SCT) matching refers to high resolution level of HLA alleles. This approach is well known and has been followed for years. As a general rule, better matching improves the overall patient and graft survival (Fig. 1).
However, this approach is also a self-limiting one. Highly alloimmunized transplant candidates whose PRA (panel reactive antibody) values exceed 85% can be successfully transplanted only with no HLA mismatches. Having said this, the possibility to find a fully matched donor for such a patient is almost zero leading to a very long waiting time for these patients.
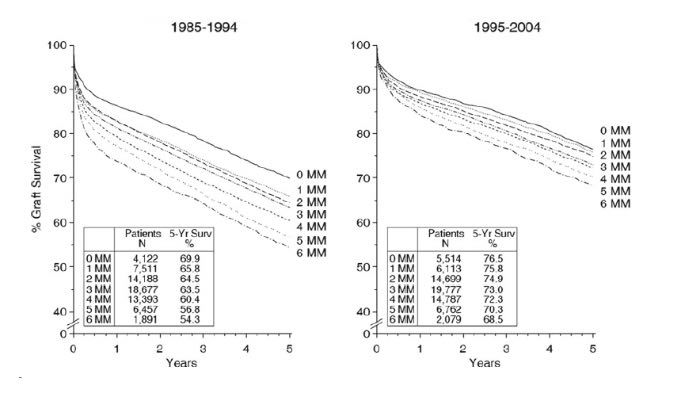
Figure 1: Graft survival rates in relation to the number of HLA-A, -B, -DR mismatches for transplants performed between 1985 and 1994 (left) and between 1995 and 2004 (right). Transplants from deceased donors were analyzed. Numbers of mismatched HLA antigens (MM) and numbers of patients studied are indicated together with graft survival rates in percent observed at 5 years post-transplant. The association of HLA matching with graft survival was statistically significant in both decades (weighted regression P < 0.001). Adopted from Süsal and Opelz (2012).
Fortunately for these patients, there is still a chance to find a suitable donor using a different matching approach. Evidence and statistics show that different criteria can be used to determine the success of HLA unmatched patients/donors transplant outcome.
In the next couple of blog posts in our series, we aim to review these matching approaches, compare them and learn more about them. In this post we will be talking about the approach used in HLA Matchmaker.
HLA Matchmaker
One of the new approaches of HLA matching was introduced in 2001 – this is matching on the epitope level. It has been developed and well described by Rene J. Duquesnoy (2001). Each HLA-specific antibody recognizes a particular structure on the target HLA molecule. This structure consists of smaller pieces called epitopes. We can look at HLA antigen as a string of epitopes represented by short sequences (triplets) involving polymorphic amino acid residues in antibody-accessible positions (Fig. 2).
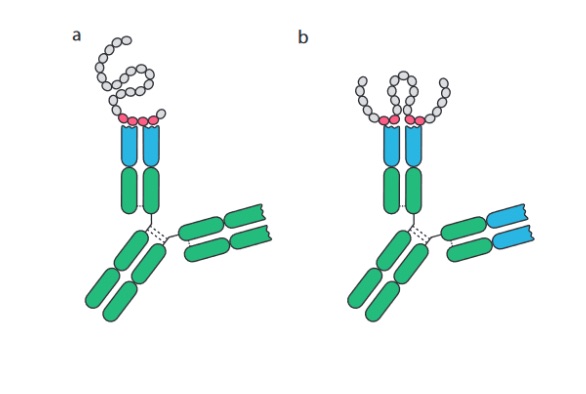
Figure 2: Schematic drawing of antibodies binding (a) linear epitopes of consecutive amino acid residues and (b) conformation or discontinuous epitopes formed by amino acids brought together by protein folding. Adopted and modified from B. Forsström (2014).
If we go into more details on antibody recognition, each antibody binding to epitopes occurs through six complementarity-determining regions (CDR) loops (Fig. 3). Three of the CDRs are on the heavy chain of the antibody molecule designated as H1, H2 and H3 and three of them are on the light chain designated as L1, L2 and L3. Each loop interacts with a small portion of amino acid residues creating a so-called structural epitope which delivers the binding strength and specificity of the antibody-antigen complex. CDR-H3 binds the centrally-located non-self amino acid residues which provide supplementary interactions that increase the stability of the antigen-antibody complex. This CDR-H3 recognized amino acid residues are so-called functional epitopes or eplets. They consist of one or more non-self amino acid residues within the patch of 3.0-3.5 Å of a given sequence position on the molecular surface. Figure 4 shows the HLA molecule from three different sites and highlights where the antibody recognizes the HLA molecule. There is a publicly accessible database of known eplets in the International HLA Epitope Registry (http://www.epregistry.com.br).
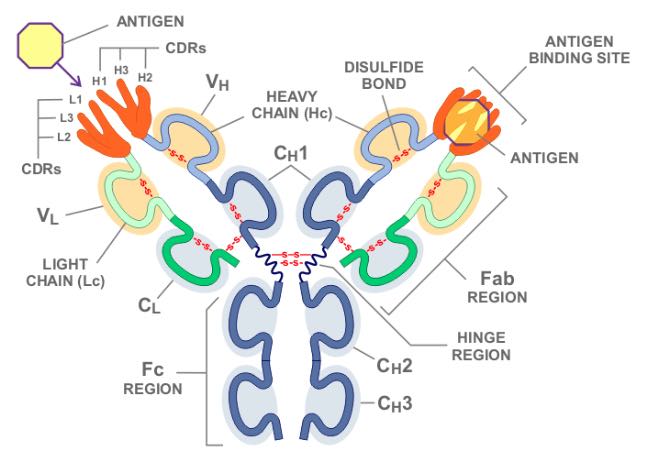
Figure 3: You can see on the left the six individual heavy and light CDR loops separately, and on the right they are combined into the full CDR1, CDR2, and CDR3. Adopted from https://biology.stackexchange.com/questions/40155/is-a-variable-domain-in-immunoglobulins-heavy-chain-different-from-the-one-in-l
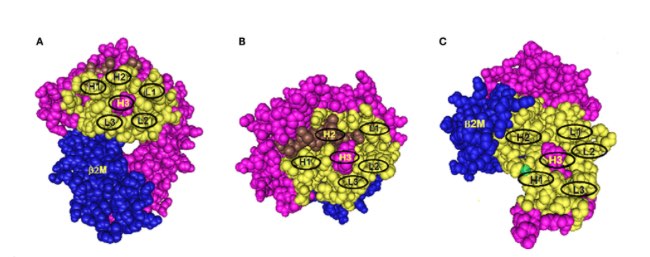
Figure 4: Three models of structural HLA class I epitope. The HLA molecule has three components: HLA chain (pink), β2-microglobulin (blue), and the bound peptide (green). The centrally located eplet (in pink) interacts with CDR-H3. Residues within a 15-Å radius are colored yellow and include configurations (in oval circles) that make contact with other CDRs on heavy chain (H1 and H2) and light chain (L1, L2, and L3). (A–C) reflect three different epitope locations on the molecule. Adopted from Duquesnoy RJ., 2016.
How is this approach translated into practical use? There have been numerous case studies showing that even patients who have received a graft from a mismatched donor have a successful transplantation outcome. Each HLA antigen has multiple epitopes that can be recognized by specific antibodies. These epitopes were characterized using three-dimensional models and despite the high polymorphism content of HLA antigens, it has become possible to determine HLA compatibility at the structural level. Imagine that each recipient has his own repertoire of self-epitopes to which no antibodies can be presented. HLA molecules share the same sequence comprising respective amino acid motifs which bring them a certain level of similarity. If the recipient shares the same motifs creating structural similarities with the donor’s antigens, then the patient will not develop HLA antibodies, or may develop a very limited number of non-harmful HLA antibodies against the donor. This approach of epitope matching, or otherwise called structural compatibility, aims to determine the number of differences between the patient and donor to assess the risk of HLA antibody development.
HLA Matchmaker is one of the algorithms that has been developed for this purpose. Its definition can be found on the official website – http://www.epitopes.net/. “HLA Matchmaker is a structurally-based computer algorithm used to determine HLA matching at the epitope level”. It focuses on epitopes recognized by HLA antibodies and the determination of epitope-based mismatch acceptability for sensitized patients considered for organ transplantation or in need of compatible platelet transfusions. In other words, it converts each HLA antigen into a string of potentially immunogenic triplets and then determines which triplets on mismatched donor HLA antigens are shared or not shared with the patient’s HLA antigens. It also explores the application of HLA mismatch permissibility at the epitope level to prevent allosensitization.
Thus, HLA Matchmaker can be used to quantify the number of mismatched eplets (triplets) to determine the risk of the mismatches. Table 1 shows an example of two HLA-B alleles – B51 (B*51:01) and B61 (B*40:02) – displaying different number of mismatched eplets with 8 different HLA phenotypes. Although in all cases we consider that as a single HLA-B mismatch, on epitope-based matching there are more differences. Firstly, B51 contains the highest number of eplet mismatch load with phenotype 1, whereas it contains no eplet mismatch load with phenotype 8. As a result, phenotype 8 would be the best candidate for transplantation.
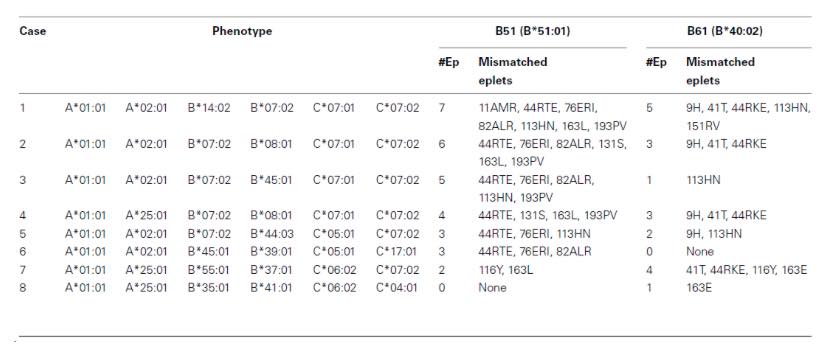
Table 1: Two examples showing how the HLA phenotype of the recipient affects the eplet load of a class I allele mismatch (Adapted from Duquesnoy R., 2011).
Examples of Structural-based Matching in Clinics
Recently, the epitope-based matching approach has been the focus of many studies and published articles show the benefit of this kind of matching. Rimando et al (2018) retrospectively tested the HLA Matchmaker algorithm on a cohort of 148 haploidentical hematopoietic stem cell transplants and observed that increased HLA epitope mismatch load was associated with a protective effect on the risk of relapse in the Graft vs Host (GVH) direction but a negative effect on time to count recovery in the Host vs Graft (HVG) direction.
A case study of Nascimento et al (2016) is also a very nice showcase of the functionality of epitope matching. This team retrospectively assessed the number of epitope mismatches between a kidney recipient and his donor after 42 years of a functioning kidney graft. The patient was haploidentical to the donor in 7 out of 8 HLA loci and identical in the DPB1 locus. At the time of the transplant, the patient developed only two donor-specific antibodies against exposed epitopes, but these did not deteriorate his condition and in the end the patient is still doing well with his 100-year old allograft.
Sullivan et al (2015) presented a comprehensive study of 4,851 pediatric heart transplantations aiming on the survival analysis for which he compared the long-term probability of graft loss by the number of eplet mismatches and allele mismatches stratified by eplet mismatches. He demonstrated that patients with lower number of eplet or allele class I mismatches had a lower risk of long-term graft loss.
Conclusion
The aforementioned case studies are only a few of the many examples that demonstrate the importance and impact of HLA matching on epitope level – currently a widely known approach for donor-recipient matching – which is also used within Eurotransplant program (among others). Although in many transplant centers the organ allocation is still based on the best antigen/allele match between patient and donor, this approach has been used as a tool for organ allocation for high-risk patients or for patients undergoing retransplantation. In current trends, we also see that this approach is gaining ground more and more in organ transplantation programs.
In our next blog post we will be discussing a different epitope-based approach and will be comparing it to HLA Matchmaker. Stay tuned…
References
- Süsal C, Opelz G. Impact of HLA matching and HLA antibodies in organ transplantation: a collaborative transplant study view. Methods Mol Biol. 2012;882:267-77.
- Duquesnoy RJ. HLAMMATCHMAKER: a molecularly based donor selection algorithm for highly alloimmunized patients. Transplant Proc. 2001 Feb-Mar;33(1-2):493-7.
- Forsström (2014) Characterization of antibody specificity using peptide array technologies, dissertation thesis, KTH Royal Institution of Technology, Stockholm, http://www.diva-portal.org/smash/get/diva2:762237/FULLTEXT01.pdf
- Duquesnoy RJ. Reflections on HLA Epitope-Based Matching for Transplantation. Front Immunol. 2016 Nov 28;7:469.
- Duquesnoy RJ. Humoral alloimmunity in transplantation: relevance of HLA epitope antigenicity and immunogenicity. Front Immunol. 2011 Nov 8;2:59
- Rimando J, Slade M, DiPersio JF, Westervelt P, Gao F, Liu C, Romee R. HLA epitope mismatch in haploidentical transplantation is associated with decreased relapse and delayed engraftment. Blood Adv. 2018 Dec 26;2(24):3590-3601.
- Nascimento E, Lucas-Junior FM, Fabreti-Oliveira RA, Vilela B, Tavora ER, Silva JP, Salomão-Filho A. Kidney Transplantation With Ultralong-Term (42 Years) Survival of a 100-Year-Old Graft. Transplant Proc. 2016 Nov;48(9):3079-3084.
- Sullivan PM, Warner P, Kemna MS, Albers EL, Law SP, Weiss NS, Law YM. HLA molecular epitope mismatching and long-term graft loss in pediatric heart transplant recipients. J Heart Lung Transplant. 2015 Jul;34(7):950-7.






