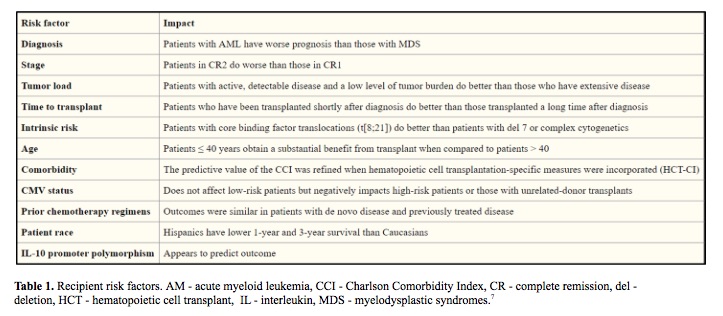
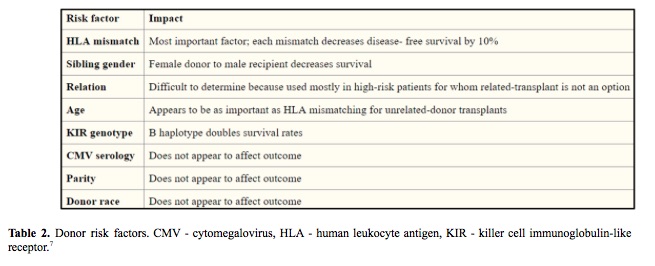
The success of hematopoietic stem cell transplant (HSCT) is influenced by many factors including HLA mismatches, variations in the major histocompatibility complex (MHC) region, minor histocompatibility targets of allorecognition, regulatory elements that affect gene expression and genetic variation that affects immune responses (Table 1, Table 2). The individual contribution from each of these factors and pathways may be challenging to quantify in individual patients, however sophisticated tools and analysis methods are available for assessing the global impact of genetic variation on transplant outcome. (1-7)
In an effort to understand and predict HSCT outcomes, we can divide the clinical features into three groups: the patient, the donor and HSCT procedure itself. Efforts to improve the success focus on optimizing modifiable factors, including better control of the patient’s disease before HSCT to lower disease recurrence, complete donor matching of HLA genes to lower the risk of graft-versus-host disease and the use of less-intense conditioning regimens to lower organ toxicity. (8-10)
A non-modifiable characteristic, that is increasingly recognized as an important factor in shaping health outcomes, is patient’s inherited genome. Its variation may affect not only predisposition to disease but also host response to disease and therapeutic interventions. (11,12) A comprehensive and systematic analysis of the clinical importance of non-HLA sequence variation within the MHC is lacking. (13,14)
As an example of an unpredictable scientific journey, full of unexpected twists and interconnections may serve a study of a role of HLA germline variation on survival after HLA-mismatched HSCT. (15)
The tested hypothesis was that inherited genome variation within the highly polymorphic MHC can impart risks to patients who undergo HSCT from HLA-mismatched donors that are not explained by patient-donor HLA mismatching.
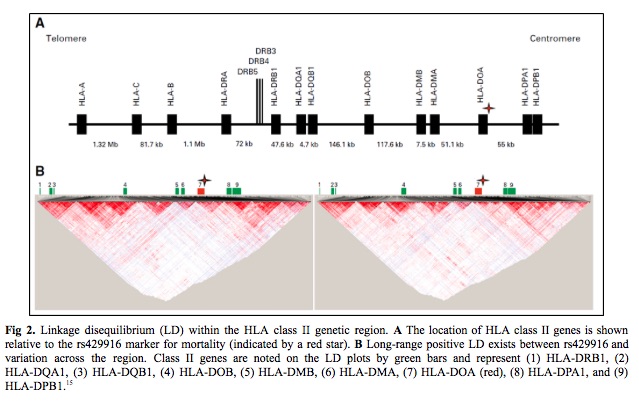
Twelve candidate transplantation determinants were identified in MHC region single nucleotide polymorphisms. (14) An association of patient rs429916 genotype with transplantation-related mortality was validated. Patients with rs429916AA and -AC genotypes had a higher 1- and 2-year transplantation-related mortality than those with rs429916CC and higher hazards of transplantation related mortality. The rs429916 genotype resides in a haplotype block that includes only HLA-DOA (Fig 2 – star). HLA-DOA encodes the DOA heterodimer of HLA-DOA, a natural inhibitor of HLA-DM involved in peptide loading of class II molecules.16,17 Three DOA proteins, DOA*01:01, DOA*01:02, and DOA*01:03 are recognized. DOA*01:01 is encoded by DOA*01:01:01 to 01:01:06 which are distinguished by silent substitutions. (18) Among known DOA alleles, only the DOA*01:01:05 is in strong positive linkage disequilibrium (LD) with rs429916A, which suggests and support rs429916A as a marker for DOA*01:01:05.
Further hypothesis to be tested was whether DOA*01:01:05 affects transplantation outcome. Increasing numbers of DOA*01:01:05 alleles were associated with higher risks of mortality and transplantation related mortality, even after adjusting for HLA mismatching. The silent substitutions that define HLA‑DOA*01:01:01 to 01:01:06 occur at sites that putatively influence methylation. It was observed that HLA-DOA expression values for DOA*01:01:05-homozygous patients are consistent with mean expression among DOA*01:01:05-negative patients. In summary, DOA*01:01:05 and rs429916A define a haplotype but neither is causative of mortality.
The next step was to determine if the true susceptibility gene is carried on DOA*01:01:05- and rs429916A positive haplotypes. The strong long-range LD between rs429916 and variants within the class II region was leveraged to identify the causal gene. The results indicated that DRB1 alleles were skewed across the entire study population and that two alleles, DRB1*03:02 and DRB1*15:03, were enriched among DOA*01:01:05-rs429916A–positive patients.
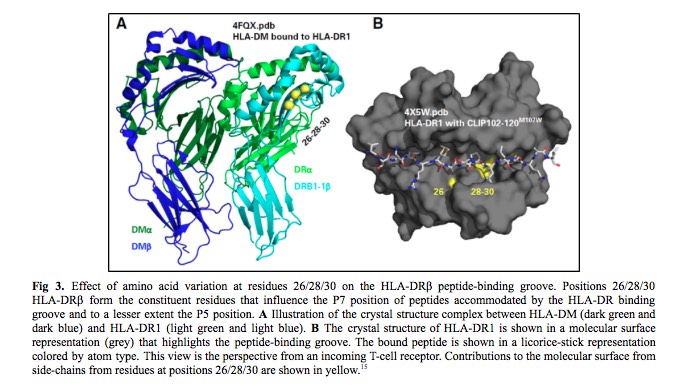
HLA nomenclature provides information on the HLA-DRB1 allele sequence, which defines the constituent amino acid residues that influence peptides accommodated by the HLA-DRβ groove (Fig 3). The third striking feature was the strong LD between HLA-DRβ residues 26/28/30 and DOA*01:01:05. DRB1*15:01 encodes FDY, DRB1*15:03 FDH, and DRB1*03:02 FEY. The frequency of FEY and FDH increased with zero, one, and two DOA*01:01:05 alleles, whereas FDY was inversely proportional. Finally, the estimated three-locus haplotype frequency of FEY-DOA*01:01:05-rs429916A in the cohort was greater than the expected frequency on the basis of the product of the three allele frequencies which supports a > 400-kb-long haplotype. In summary, rs429916A-positive patients encode different HLA-DRβ residues than rs429916A-negative patients.
Evaluation of residues shared among distinct HLA-DRB1 alleles may shed light on common features that are biologically important in transplantation survivorship. This is why the last task in this story was to test the hypothesis that mortality depends on the presence of specific amino acid substitutions in the peptide-binding region of HLA-DRβ. HLA-DRB1 alleles that encode the FEY motif at residues 26/28/30 of HLA-DRβ in patients were determined to increase the risk of mortality after HLA-mismatched unrelated donor transplantation.
Taken together, the risk of mortality after transplantation is influenced by HLA-DRB1 alleles with substitutions that influence the peptide binding region of HLA-DR molecules. The study suggested that determination of patients with high-risk HLA-DRB1 alleles will enhance risk assessment.
Proving that the risk of mortality associated with HLA-DR/DOA did not depend on the kind of HLA mismatch, nor was it a consequence of more HLA-DRB1 mismatching among patients of different origin, the study might be leading us to a wide field of not necessarily only HLA genes impact to be discovered. Let’s see how many clinically important heritable variants are just waiting to be identified to extend our understanding of the genetics of transplantation!
References:
- Lee SJ, Klein J, Haagenson M, et al. High resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13): 4576-4583.
- Petersdorf EW. The major histocompatibility complex: a model for understanding graft-versus-host disease. Blood. 2013;122(11):1863-72.
- Trowsdale J. HLA genomics in the third millennium. [Review] Curr Opin Immunol. 2005; 17(5):498-504
- Ali JM, Bolton EM, Bradley JA, Pettigrew GJ. Allorecognition pathways in transplant rejection and tolerance. Transplantation 2013;96:681-8.
- Middleton PG, Cullup H, Dickinson AM, et al. Vitamin D receptor gene polymorphism associates with graft-versus-host disease and survival in HLA-matched sibling allogeneic bone marrow transplantation. Bone Marrow Transplant. 2002;30(4): 223-228.
- Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. [Review] J Hum Genet. 2009;54(1):15-39
- Anasetti C. What are the most important donor and recipient factors affecting the outcome of related and unrelated allogeneic transplantation?. Best Pract Res Clin Haematol. 2008;21(4):691-7.
- Kollman C, Spellman SR, Zhang MJ, et al: The effect of donor characteristics on survival after unrelated donor transplantation for hematologic malignancy. Blood 127:260-267, 2016
- Petersdorf EW, Malkki M, O’hUigin C, et al: High HLA-DP expression and graft-versus-host disease. N Engl J Med 373:599-609, 2015
- Storb R, Sandmaier BM: Nonmyeloablative allogeneic hematopoietic cell transplantation. Haematologica 101:521-530, 2016
- Weinshilboum R: Inheritance and drug response. N Engl J Med 348:529-537, 2003
- Chien JW, Zhang XC, Fan W, et al: Evaluation of published single nucleotide polymorphisms associated with acute GVHD. Blood 119:5311-5319, 2012
- Sato-Otsubo A, Nannya Y, Kashiwase K, et al: Genome-wide surveillance of mismatched alleles for graft-versus-host disease in stem cell transplantation. Blood 126:2752-2763, 2015
- Petersdorf EW, Malkki M, Horowitz MM, et al: Mapping MHC haplotype effects in unrelated donor hematopoietic cell transplantation. Blood 121: 1896-1905, 2013
- Petersdorf EW, et al: Patient HLA Germline Variation and Transplant Survivorship. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 36 24 (2018): 2524-2531
- Denzin LK: Inhibition of HLA-DM mediated MHC class II peptide loading by HLA-DO promotes self tolerance. Front Immunol 4:465, 2013
- Mellins ED, Stern LJ: HLA-DM and HLA-DO, key regulators of MHC-II processing and presentation. Curr Opin Immunol 26:115-122, 2014
- The IPD-IMGT/HLA Database: Welcome to IPD-IMGT/HLA. https://www.ebi.ac.uk/ipd/imgt/hla
Article written by Vera Siffnerova (FAS, Omixon)