After last weeks post about randomly subsampling fastq files, let’s take a look at a different method for making read files smaller.
A very easy method, that can be used for 454 or IonTorrent data (but usually not for Illumina) is getting rid of very short reads. If you take a look at the quality control results of the IonTorrent example file, you can see, that read length range is between 5 and 399, while most reads are around 200 bases long. As an exercise, let’s get rid of reads shorter than 50 bases.
A possible solution for this is the “fastx_clipper” command of the FastX toolkit:
fastx_clipper -Q33 -l 50 -n -v -i SRR515927.fastq -o SRR515927_mt50len.fastq
It’s always a goof policy to give descriptive filenames (hence the mt50len part). Believe me, two months from now, you’ll have absolutely no idea what SRR515921_filtered.fastq contains exactly.
You can also filter by length using for example Prinseq. To mix it up a little, let’s filter out all reads that are shorter than 50 or longer than 350 bases:
perl prinseq-lite.pl -fastq SRR515927.fastq -min_len 50 -max_len 350 -out_good SRR515927_mt50_lt350len
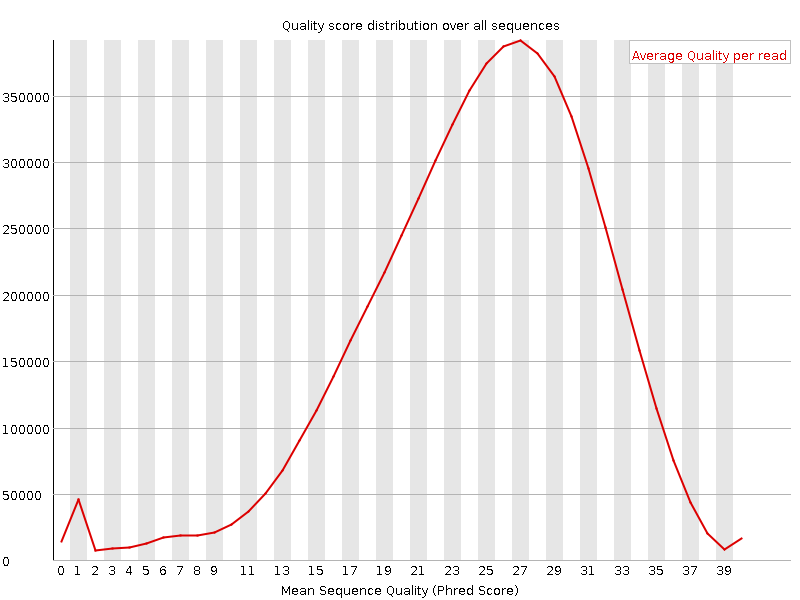
Let’s try some quality filtering as well! You might remember, that the Illumina example data set has very low quality:
Let’s filter out all reads with less than 10 average quality!
I will use Prinseq again, for two reasons: it has surprisingly diverse filtering options (e.g. GC-content based filtering) and it can deal with paired reads in a single step, so it saves us the additional step of fixing read pairs (you can use e.g. Biopieces for that).
perl prinseq-lite.pl -fastq SRR022913_1.fastq -fastq2 SRR022913_2.fastq -min_qual_score 10 -out_good SRR022913_mt10qual
If we take a look at the produced files, we can see that two filtered and two singleton files were produced and the “non-singleton” files have the exact same length:
$ wc -l SRR022913* 24818636 SRR022913_1.fastq 24818636 SRR022913_2.fastq 362100 SRR022913_mt10qual_1.fastq 1410188 SRR022913_mt10qual_1_singletons.fastq 362100 SRR022913_mt10qual_2.fastq 770604 SRR022913_mt10qual_2_singletons.fastq