After a few weeks long sidetrack about reference sequence manipulation, let’s get back to our reads. If you’ve read my post about read quality control results, you might remember, that I found some untrimmed adaptors on the 454 reads (SRR797242.fastq). In this post, I’ll show you a few ways to remove these adaptors from the reads.
First, let’s take a quick look at the FastQC result again!
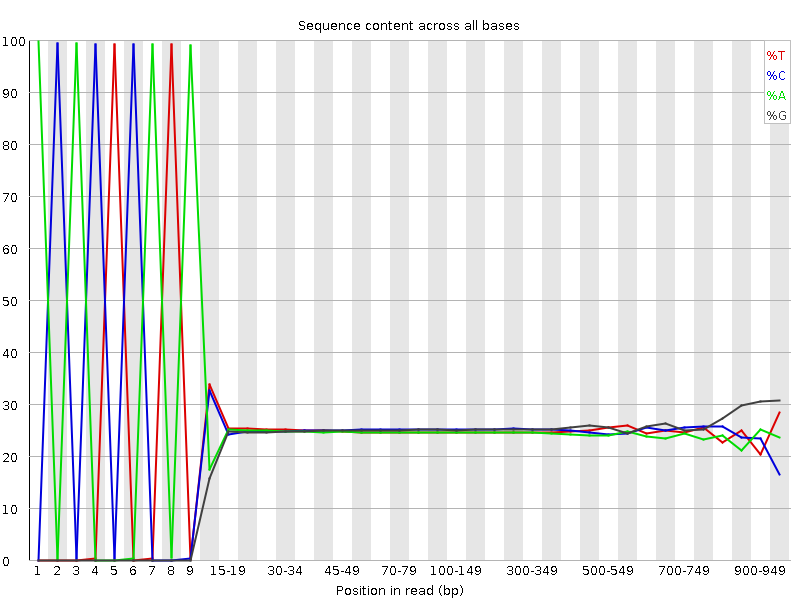
Per base sequence content of the 454 read file. By “reading” the diagram, you can figure out the sequence of the adaptor. We have green-blue-green-blue-red-blue-green-red-green, which is equevalent to ACACTCATA.
A very simple way to check, whether this is actually the adaptor or not, is the ‘grep’ command, which is a string search tool, available in Linux/Unix. The search syntax is the following:
grep '^ACACTCATA' SRR797242.fastq #The "^" character before the string we're searching for tells grep... #...to search only at the beginning of the lines in the file. #Alternatively, you can use the following syntax: cat SRR797242.fastq | grep '^ACACTCATA'
By default, grep outputs all the lines that contain the specified string, so the beginning of the output should look something like this:
$ grep '^ACACTCATA' SRR797242.fastq | head ACACTCATACTCATCTGCGGATCATGCTAATCGCCGCCTGACAATTATTCACTCAAGGCACACAGGGAGTA ACACTCATACTGCAACGGAGTCTGCGAGGACGCTTCCTGAATCGTCTTATTGCAGTGAATGACAGGCAATGCGGAAGCAGCTACGCAAACGCAACAACTTTGCGCAAAGTGTGAGCAAGGGCTACGTCACATGGCCGCGCCGTGTATAATAAGCTCGTATGTAGGCTTTATTCGCTAATCACATACGAAGATACTCATGGCTCAAGGCACACAGGGGATA ACACTCATACTCCGATTCGTGCTTGATACGTTCTGCGGTGGCTTCACCGATCAGAGAACCGTAATTACGACGCACATAGTTGATGATAGCTTCGTCGAAACGGTCACCACCAAGGCACA ACACTCATACTCCGATTCGTGCTTGATACGTTCTGCGGTGGCTTCACCGATCAGAGACCGTATTACGACGCACATAGTTGATGATAGCTTCGTCGAACGGTCACCACCAAGGCACACAGGGATA ACACTCATACTCCAGCGAGCGCGGGCACTCACGAGAAGCAGTGATGGACTCAGTAGTGCGTTCAATGGAAGACTATATCAACTACATCACACCGCAGTTTTCCCGCACCACT ACACTCATACTGGTGACCAGCACTTCACCGGGCAACAAGGTGCGGGCAAATGCCCTGATAACGTGCCTGCCGCGCCTGGGCCTCAATGCCCAGTCCTTCTTGCGCAAGTTGTACGCGTTCGACCACCAGCGGCACCTTGCCACTGTTAGTA ACACTCATACTGCTGGATTACACTCCGCGTGAGTCGTTCCTGAAT ACACTCATACTACTGGTGGTATGGCAACGGACATGTTATGGCGC ACACTCATACTTTACAGCTGTTGCGCGAACTGCAAGGCGACGACTGGGAATAAGTGGCATGCTGTTTATTACTCATAACCTCAGCATTGTCAGAAAACTGGCCCACCGCGTGGCGGTAATGCAAAACGGTCGCTGTGTCGAGCAAAATTACGCCGCTACGCTATTTGCATCACCCACTCATCCTTACACACAAAAGCTACTCAACAGTGAACCGTCAGGCGATCCAGTGCCGTTGCC ACACTCATACTGCTGGATTACACTCCGCGTGAGTCGTTCCTGAATCCGGGT
To count the lines starting with the adaptor sequence, you can use the “-c” flag:
$ grep -c '^ACACTCATA' SRR797242.fastq 1370973
This suppresses the normal output and only the total number of lines matching the search expression are printed.
Now, to compare this number to the total number of reads, without digging up the quality control results, we can check the total number of read IDs in the fastq file in the terminal.
Similarly as before, we can use grep:
$ grep -c '^@' SRR797242.fastq 1387255
Of course, as Linux is “the land of the free”, you can achieve the same thing in at least a dozen different ways. For example with awk and wc (it’s an abbreviation of word count, not water closet).
awk '/^@/' SRR797242.fastq | wc -l 1387255
As you can see, the adaptor can be found at the beginning of basically all the reads. As we were only searching for exact matches, it is quite possible, that the rest of the reads has the adaptor as well, they just have some sequencing error(s) in the adaptor region of the read.
I will show you a few different tools, but feel free to share additional tools, scripts and opinions in the comment section!
Let’s start with ea-utils! The syntax is the following:
$fastq-mcf adapter.fa SRR797242.fastq -o SRR797242_eautils.fastq Scale used: 2.2 Phred: 33 Threshold used: 751 out of 300000 Adapter adapter_454 (ACACTCATA): counted 297041 at the 'start' of 'SRR797242.fastq', clip set to 1 Files: 1 Total reads: 1385764 Too short after clip: 2 Clipped 'start' reads: Count: 1385528, Mean: 8.93, Sd: 0.92
As you can see from the output, some statistics about the run are automatically generated. Also, reads that are too short after clipping the adaptor are filtered out. To validate the success of the adaptor removal, we can run the grep command again with the new file:
$grep -c '^ACACTCATA' SRR797242_eautils.fastq 0
Biopieces is another possible tool for this task. This bioinformatics suite uses a slightly more complicated syntax, but it contains several very useful tools.
$read_fastq -i SRR797242.fastq | find_adaptor -f ACACTCATA | clip_adaptor | write_fastq -o SRR797242_biopieces.fastq -x $grep -c '^ACACTCATA' SRR797242_biopieces.fastq 0
There are several other tools available for adapter removal (e.g. take a look at this list on Bioscholar.com or this SeqAnswers thread.)






