I recently ran into an email on one of the numerous mailing lists I’m subscribed to. The email was written by a student who was desperately looking for NGS data for testing a pipeline. This letter made me realise that finding short read data is probably not an easy task for people who are just starting to use next generation sequencing techniques. If you know the right search phrases, you can easily find the short read databases, but if you’re googling “NGS data” you’ll get basically no relevant hits. So here is a collection of NGS data resources to make the life of the newbies a little easier.
You can find NGS reads at exactly the same webpages where you can find Sanger data:
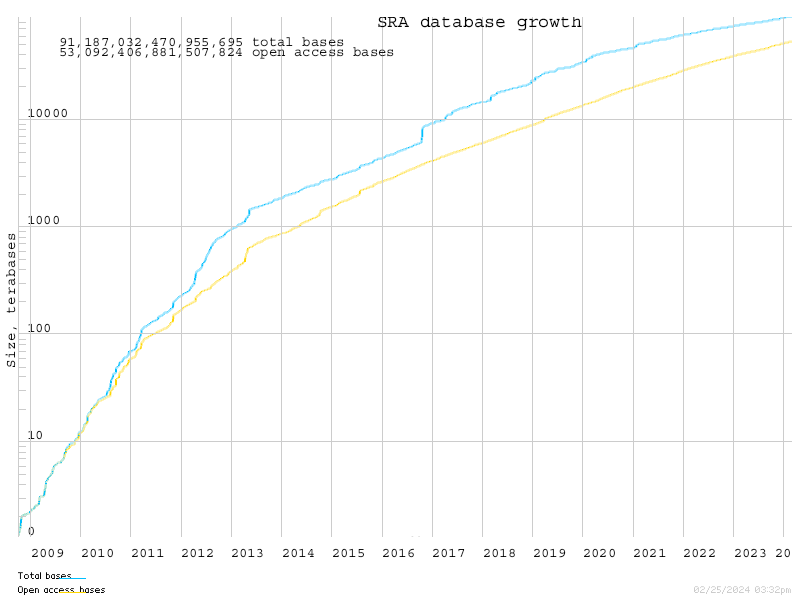
The number of short read datasets available from the NCBI SRA skyrocketed in the last few years.
(Source: http://www.ncbi.nlm.nih.gov/Traces/sra/i/g.png)
It’s usually a good idea to take a look around the webpages of next generation sequencing companies, they generally provide some example data sets as well. For example:
If you are interested in human data, you should take a look at the 1000 Genomes project.
Also, a few biological database collections:
