We all need a little reassurance sometimes…
In the past few years Next Generation Sequencing has conquered immunology labs all around the world and is actively used in both research and diagnostics. But how was it possible to develop and validate these methods? How do we verify the accuracy of our assay, the Holotype HLA kit and the HLA Twin software? Well, with the help of typing buddies of course.
As you could see in our previous blogpost (Genotyping Engine Race – Measuring Performance) we use huge datasets for validation purposes, which were previously genotyped by either of the following methods.
The Terminator
Good ol’ Sanger sequencing is based on using chain-terminating dideoxynucleotides in order to create fragments of different lengths. These fragments can be identified by labelling the primer, the dNTPs or most commonly the ddNTPs with radioactive tags or fluorescent dye, and separating them by size using gel electrophoresis. Automated DNA-sequencing instruments have built-in capillary electrophoresis, can detect fluorescence, and output data as fluorescent peak trace chromatograms, however the long range PCR amplification of the targeted regions still need to be done independently. A nice invention from the 2000’s uses lab-on-a-chip technology (microfluidic Sanger sequencing), in which the sequencing steps are integrated on a chip using nanoliter-scale sample volumes.
Sanger sequencing is considered the “gold standard” typing method because of its accuracy and resolution, however it’s more expensive and less efficient compared to the other methods.
Technical requirements: PCR and primers for targeted genes, DNA sequencer, labelled ddNTP-s
Resolution: high (4-field)
Time: 1-2 days
The probe-ability of polymorphisms
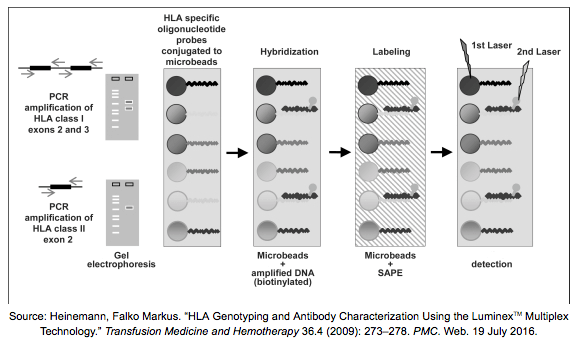
It’s not always necessary to sequence the samples when looking for genotypes, we can concentrate on the individual polymorphisms that distinguish alleles from each other. In a method called PCR-SSO (Sequence Specific Oligonucleotides) we amplify our targeted regions with primers that are labelled with Biotin. The amplicons are hybridized to sequence specific probes attached to microbeads and SAPE (streptavidin phycoerythrin) is added to the mixture – these molecules bind to Biotin and can emit fluorescence. The surface of these beads are colored uniquely with a mix of two infrared dyes, therefore with the help of flow cytometry it is possible to detect the beads that are covered with our bound targets and determine the genotypes. Probes are also helpful in antibody screening and can be implemented on microarrays as well.
SSO assays are easy and convenient, but they only cover a narrow set of genotypes.
Technical requirements: SSO reagents, PCR, electrophoresis, flow cytometry analyzer
Resolution: from low to intermediate (usually 2-field)
Time: 4-6 hours
Primers – unique, specific, marvellous!
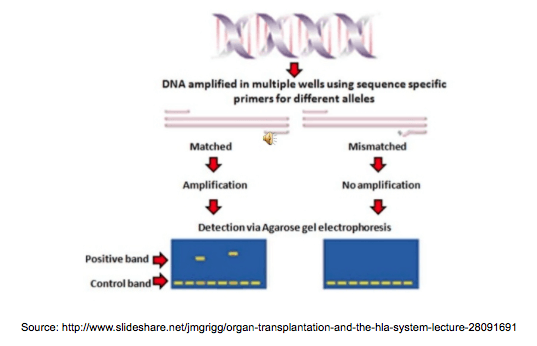
Working with primers – that rings a bell, huh? Right, we use primers in all of the technologies listed above (in NGS too!), however the PCR-SSP (Sequence Specific Primers) method has a very clever way of using their main feature: sequence specificity. We can find assays for HLA loci, KIR and a wide range of disease associated genes, each of them containing unique mixes of primers, which amplify specific regions in different alleles. They also include positive and negative controls, so we can make sure our assay works as it’s supposed to. So all we need to do is add our sample, put the assay in a PCR, then run the amplicons on a gel – the genotypes can be easily identified by the pattern of the amplicons.
SSP is an easy, quick and reliable construction, however the diversity of the alleles and the size of the assay limit the obtainable resolution.
Technical requirements: SSP assay, PCR, gel electrophoresis
Resolution: depends on the assay, from intermediate to high (usually 3-field)
Time: 3 hours
Although we’ve summed up that each of our typing buddies has its own ups and downs, let’s not forget that we can always rely on them when in doubt!
Science is fun!
/ by Tünde Lilla Vágó